Loki2 in silico Immunostaining
This notebook demonstrates how to prepare CRC02 patch-level data, train a LightGBM regressor on HE embeddings to predict IF channel intensity, and visualize predictions.
Data Requirements
The example data is stored in the directory ../data/imm_stain, which can be donwloaded from Google Drive.
You will need:
Loki2 cell inference results
Paired HE and IF patches
import os
from pathlib import Path
import json
import numpy as np
import torch
import matplotlib.pyplot as plt
import loki2.immstain
# Display inline plots
%matplotlib inline
Examples of PanCK Marker
Configure paths and parameters
Set your dataset locations and model options. Update these to match your filesystem.
cfg = loki2.immstain.TutorialConfig(
he_ome_path=Path("../data/imm_stain/CRC02-HE.ome.tif"),
global_nuclei_json=Path("../data/imm_stain/CRC02-HE_PYRAMIDAL_cells.json"),
global_embedding_pt=Path("../data/imm_stain/CRC02-HE_PYRAMIDAL_cells.pt"),
if_patch_dir=Path("../data/imm_stain/patches_IF_lvl0_2048_sel"),
if_patch_pattern="IF_sel_r{row:03d}_c{col:03d}.tif",
patch_json_dir=Path("../outputs/imm_stain/CRC02_patch_cells_json"),
patch_emb_dir=Path("../outputs/imm_stain/CRC02_patch_embeddings"),
level0_idx=0,
level1_idx=1,
patch_size=2048,
)
# Training choices
target_channel = 4 # example: Pan-CK channel (0-based index in IF stack)
use_log = True # log1p on targets
normalize_embeddings = True
extend_nuclei = True # watershed grow nuclei to cell boundaries
expansion_distance = 5 # pixels for watershed limit
# Paths to save outputs
model_out_path = Path(f"../outputs/imm_stain/models/CRC02_lightgbm_channel{target_channel}.pt".format(ch=target_channel))
train_pred_path = Path(f"../outputs/imm_stain/predictions/CRC02_channel{target_channel}_train_predictions.json".format(ch=target_channel))
test_pred_path = Path(f"../outputs/imm_stain/predictions/CRC02_channel{target_channel}_test_predictions.json".format(ch=target_channel))
One-time prep: generate per-patch JSON and embeddings
Step 1: Split global nuclei + embeddings into per-patch JSON/PT files (one-time prep)
Run this once to split the global nuclei JSON + embeddings into per-patch files. Skip if you already have CRC02_patch_cells_json/ and CRC02_patch_embeddings/ populated.
do_generate = True # set True for first run
if do_generate:
loki2.immstain.generate_patch_json_and_embeddings(cfg)
else:
print("Skipping generation; set do_generate=True to run.")
Loading global embeddings...
Global embeddings: 1038158 cells, dim=1280
Loading global nucleus JSON...
../data/imm_stain/CRC02-HE.ome.tif level 0 zarr shape: (3, 54363, 82230)
../data/imm_stain/CRC02-HE.ome.tif level 1 zarr shape: (3, 27182, 41115)
Level0: H0=54363, W0=82230
Level1: H1=27182, W1=41115
Scale factors: scale_y=0.500009, scale_x=0.500000
Patch grid (level1): 14 rows x 21 cols
Assigning global cells to patches (level1 grid)...
Non-empty patches: 188
Saving patch JSON + embedding files...
Done generating patch-specific JSON + embeddings.
Inspect available patches
List patches that have both nuclei/embedding JSON and IF patch present.
available_patches = loki2.immstain.list_available_patches(cfg)
print(f"Found {len(available_patches)} patches with both JSON/emb and IF:")
print(available_patches) # show a sample
# use those patches for quick testing
available_patches=['r004_c006','r004_c005','r004_c007','r003_c006']
Found 4 patches with both JSON/emb and IF:
['r003_c006', 'r004_c005', 'r004_c006', 'r004_c007']
Define train/val/test patch splits
Select a validation patch (held out from train) and optional test patches for final evaluation/visualization.
# Simple deterministic split example (edit as needed)
if len(available_patches) < 3:
raise RuntimeError("Need at least 3 patches to run train/val/test example.")
val_patch = available_patches[0]
train_patches = available_patches[1:-1]
test_patches = [available_patches[-1]]
print("Validation patch:", val_patch)
print("Train patches (count):", len(train_patches))
print("Test patches:", test_patches)
Validation patch: r004_c006
Train patches (count): 2
Test patches: ['r003_c006']
Build datasets
Step 2: Build train/val/test cell-level datasets for a chosen IF channel
Creates cell-level feature matrix X, targets y, and metadata for the chosen IF channel.
with loki2.immstain.timed_section(f"build train dataset (channel {target_channel})"):
X_train, y_train, meta_train = loki2.immstain.build_dataset_for_patches(
cfg,
train_patches,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei
)
with loki2.immstain.timed_section(f"build val dataset (channel {target_channel})"):
X_val, y_val, meta_val = loki2.immstain.build_dataset_for_patches(
cfg,
[val_patch],
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei
)
print("Train shape", X_train.shape, "Val shape", X_val.shape)
Processing patch r004_c005...
N nuclei in patch JSON: 16911
Patch embeddings shape: (16911, 1280)
Processing patch r004_c007...
N nuclei in patch JSON: 16068
Patch embeddings shape: (16068, 1280)
Total cells across all patches: 32979, embedding dim=1280
[timer] build train dataset (channel 4): 146.89s
Processing patch r004_c006...
N nuclei in patch JSON: 15974
Patch embeddings shape: (15974, 1280)
Total cells across all patches: 15974, embedding dim=1280
[timer] build val dataset (channel 4): 48.28s
Train shape (32979, 1280) Val shape (15974, 1280)
Preprocess features and targets
Optionally L2-normalize embeddings and log-transform targets (log1p).
def prep_features(X):
if not normalize_embeddings:
return X.copy()
return loki2.immstain.l2_normalize_embeddings(X.copy())
prep_target = (np.log1p if use_log else (lambda x: x.copy()))
X_train_proc = prep_features(X_train)
X_val_proc = prep_features(X_val)
y_train_proc = prep_target(y_train)
y_val_proc = prep_target(y_val)
print("Preprocessed shapes:", X_train_proc.shape, X_val_proc.shape)
Preprocessed shapes: (32979, 1280) (15974, 1280)
Train LightGBM with patch-level validation
Step 3: Train a LightGBM regressor with per-patch holdout validation
Only the LightGBM model is used to keep the pipeline focused.
with loki2.immstain.timed_section("train lightgbm"):
model, val_metrics, y_val_pred = loki2.immstain.train_lightgbm_regressor(
X_train_proc,
y_train_proc,
X_val_proc,
y_val_proc,
random_state=42,
num_boost_round=2000,
early_stopping_rounds=100,
)
print("Validation metrics:", json.dumps(val_metrics, indent=2))
# Train predictions (for diagnostics on processed scale)
y_train_pred = loki2.immstain.predict_with_model(model, X_train_proc)
Training until validation scores don't improve for 100 rounds
[100] train's rmse: 0.603496 valid's rmse: 0.829716
[200] train's rmse: 0.48877 valid's rmse: 0.821889
[300] train's rmse: 0.405244 valid's rmse: 0.819101
[400] train's rmse: 0.340861 valid's rmse: 0.817872
[500] train's rmse: 0.289398 valid's rmse: 0.817132
[600] train's rmse: 0.247122 valid's rmse: 0.816637
[700] train's rmse: 0.211582 valid's rmse: 0.816496
[800] train's rmse: 0.182258 valid's rmse: 0.816471
Early stopping, best iteration is:
[720] train's rmse: 0.205237 valid's rmse: 0.81629
[timer] train lightgbm: 2431.94s
Validation metrics: {
"mse": 0.6663286154146737,
"mae": 0.6359814654748486,
"r2": 0.3973233849316552,
"pearson_r": 0.6339607099617551,
"ssim": 0.3455918097967648,
"model_type": "lightgbm"
}
Optional: test set evaluation
Evaluate on held-out patches (if provided) and collect predictions.
test_results = None
if test_patches:
with loki2.immstain.timed_section(f"build test dataset (channel {target_channel})"):
X_test, y_test, meta_test = loki2.immstain.build_dataset_for_patches(
cfg,
test_patches,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei
)
X_test_proc = prep_features(X_test)
y_test_proc = prep_target(y_test)
with loki2.immstain.timed_section("predict test"):
y_test_pred = loki2.immstain.predict_with_model(model, X_test_proc)
if use_log:
y_test_eval = y_test
y_test_pred_eval = np.expm1(y_test_pred)
else:
y_test_eval = y_test
y_test_pred_eval = y_test_pred
test_metrics = loki2.immstain.compute_regression_metrics(y_test_eval, y_test_pred_eval)
print("Test metrics (original scale):", json.dumps(test_metrics, indent=2))
test_results = {
"X": X_test_proc,
"y_true": y_test_proc,
"y_pred": y_test_pred,
"meta": meta_test,
"metrics": test_metrics,
"y_true_raw": y_test_eval,
"y_pred_raw": y_test_pred_eval,
}
else:
print("No test patches provided; skip test evaluation.")
Processing patch r003_c006...
N nuclei in patch JSON: 14871
Patch embeddings shape: (14871, 1280)
Total cells across all patches: 14871, embedding dim=1280
[timer] build test dataset (channel 4): 59.56s
[timer] predict test: 0.08s
Test metrics (original scale): {
"mse": 8509761.026050935,
"mae": 1978.1806053469938,
"r2": 0.059515598904488654,
"pearson_r": 0.39398436519666197,
"ssim": 0.30092517473176844
}
Save model and predictions
Step 4: Save per-cell predictions to JSON and plot diagnostics (pred vs. true, residuals)
Persist the trained model plus per-cell prediction JSON for train/val/test splits.
# Save model bundle
model_out_path.parent.mkdir(parents=True, exist_ok=True)
torch.save(
{
"model": model,
"normalize_embeddings": normalize_embeddings,
"use_log": use_log,
"metrics": val_metrics,
},
model_out_path,
)
print(f"Saved model to {model_out_path}")
# Save train predictions (processed scale)
loki2.immstain.save_prediction_json(
train_pred_path,
channel_idx=target_channel,
extend_nuclei=extend_nuclei,
normalize_embeddings=normalize_embeddings,
metrics=val_metrics,
meta=meta_train,
y_true=y_train_proc,
y_pred=y_train_pred,
)
# Save test predictions if available
loki2.immstain.save_prediction_json(
test_pred_path,
channel_idx=target_channel,
extend_nuclei=extend_nuclei,
normalize_embeddings=normalize_embeddings,
metrics=(test_results or {}).get("metrics", val_metrics),
meta=(test_results or {}).get("meta", []),
y_true=(test_results or {}).get("y_true", []),
y_pred=(test_results or {}).get("y_pred", []),
)
Saved model to ../outputs/imm_stain/models/CRC02_lightgbm_channel4.pt
Per-cell predictions saved to: ../outputs/imm_stain/predictions/CRC02_channel4_train_predictions.json
Per-cell predictions saved to: ../outputs/imm_stain/predictions/CRC02_channel4_test_predictions.json
Visualize predictions
Quick scatter and residual plots on validation predictions (processed scale). For raw-scale plots, invert log1p before plotting.
Patch-level GT vs Pred visualization
Render one patch with per-cell measured IF intensity, predicted intensity, residual heatmap, and scatter. Uses the saved prediction JSON.
patch_for_viz = 'r004_c007'
pred_json_for_viz = train_pred_path
out_path = Path("../outputs/imm_stain") / "figures_viz_only" / f"patch_{patch_for_viz}_ch{target_channel}_tutorial.png"
fig, axes = loki2.immstain.visualize_patch_prediction(
cfg,
patch_name=patch_for_viz,
prediction_json_path=pred_json_for_viz,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei,
predictions_are_log=use_log,
colorbar_range=None,
save_path=out_path
)
plt.show()
print(f"Saved patch viz to {out_path}")
Saved patch visualization to ../outputs/imm_stain/figures_viz_only/patch_r004_c007_ch4_tutorial.png
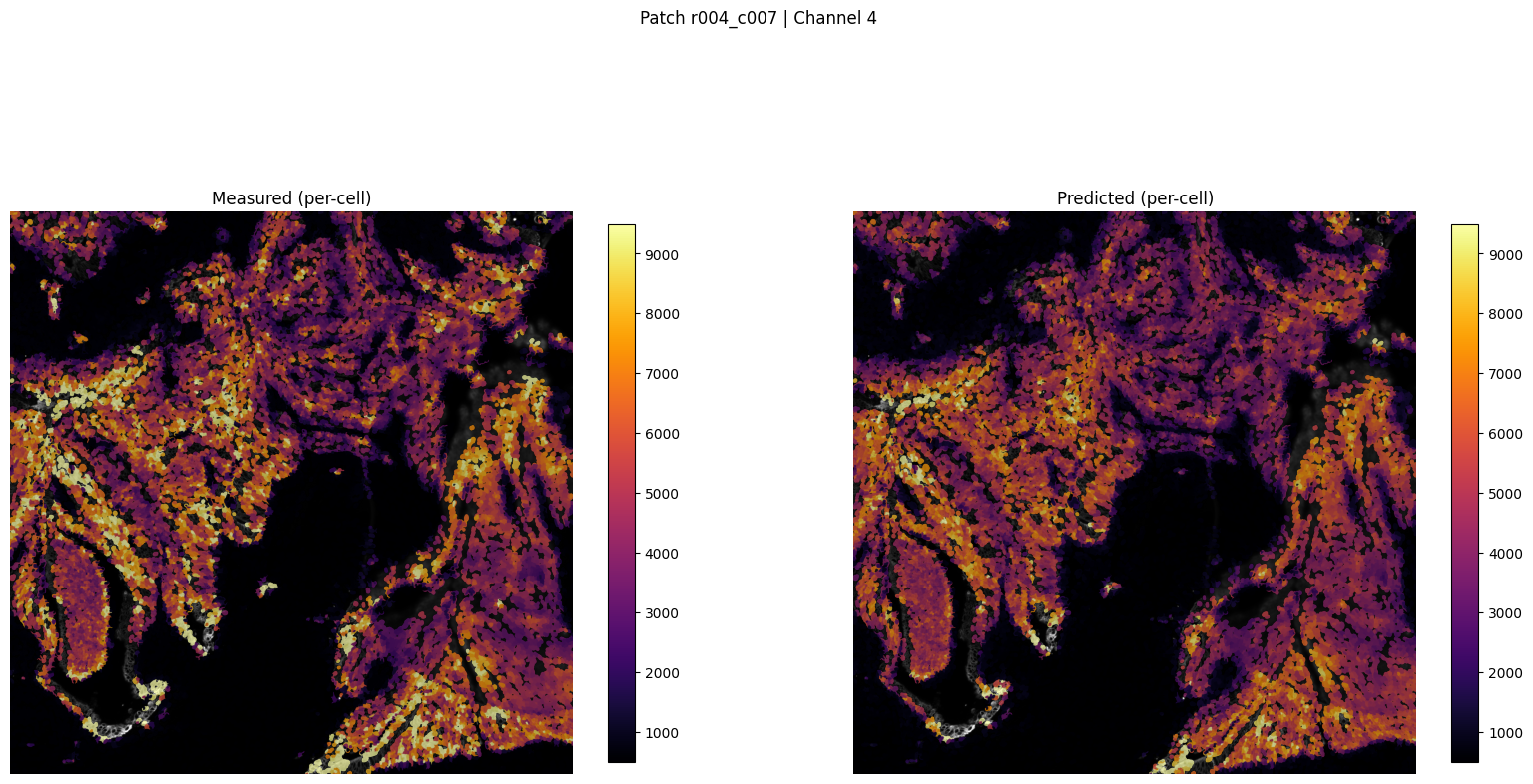
Saved patch viz to ../outputs/imm_stain/figures_viz_only/patch_r004_c007_ch4_tutorial.png
patch_for_viz = test_patches[0] if test_patches else val_patch
pred_json_for_viz = test_pred_path if test_patches else train_pred_path
out_path = Path("../outputs/imm_stain") / "figures_viz_only" / f"patch_{patch_for_viz}_ch{target_channel}_tutorial.png"
fig, axes = loki2.immstain.visualize_patch_prediction(
cfg,
patch_name=patch_for_viz,
prediction_json_path=pred_json_for_viz,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei,
predictions_are_log=use_log,
colorbar_range=None,
save_path=out_path
)
plt.show()
print(f"Saved patch viz to {out_path}")
Saved patch visualization to ../outputs/imm_stain/figures_viz_only/patch_r003_c006_ch4_tutorial.png
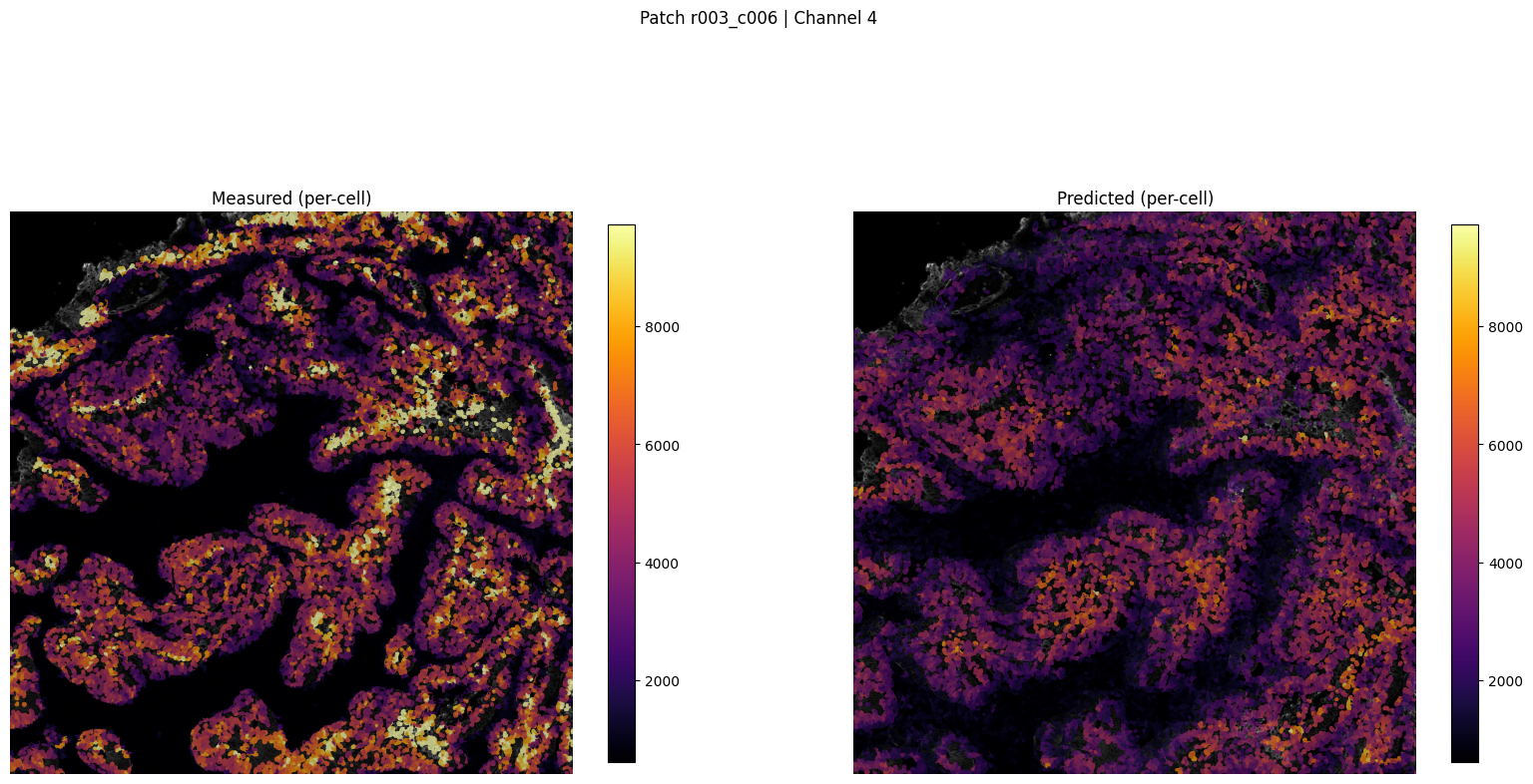
Saved patch viz to ../outputs/imm_stain/figures_viz_only/patch_r003_c006_ch4_tutorial.png
Examples of CD45RO Marker
Configure paths and parameters
Set your dataset locations and model options. Update these to match your filesystem.
cfg = loki2.immstain.TutorialConfig(
he_ome_path=Path("../data/imm_stain/CRC02-HE.ome.tif"),
global_nuclei_json=Path("../data/imm_stain/CRC02-HE_PYRAMIDAL_cells.json"),
global_embedding_pt=Path("../data/imm_stain/CRC02-HE_PYRAMIDAL_cells.pt"),
if_patch_dir=Path("../data/imm_stain/patches_IF_lvl0_2048_sel"),
if_patch_pattern="IF_sel_r{row:03d}_c{col:03d}.tif",
patch_json_dir=Path("../outputs/imm_stain/CRC02_patch_cells_json"),
patch_emb_dir=Path("../outputs/imm_stain/CRC02_patch_embeddings"),
level0_idx=0,
level1_idx=1,
patch_size=2048,
)
# Training choices
target_channel = 2
use_log = True # log1p on targets
normalize_embeddings = True
extend_nuclei = True # watershed grow nuclei to cell boundaries
expansion_distance = 5 # pixels for watershed limit
# Paths to save outputs
model_out_path = Path(f"../outputs/imm_stain/models/CRC02_lightgbm_channel{target_channel}.pt".format(ch=target_channel))
train_pred_path = Path(f"../outputs/imm_stain/predictions/CRC02_channel{target_channel}_train_predictions.json".format(ch=target_channel))
test_pred_path = Path(f"../outputs/imm_stain/predictions/CRC02_channel{target_channel}_test_predictions.json".format(ch=target_channel))
One-time prep: generate per-patch JSON and embeddings
Step 1: Split global nuclei + embeddings into per-patch JSON/PT files (one-time prep)
Run this once to split the global nuclei JSON + embeddings into per-patch files. Skip if you already have CRC02_patch_cells_json/ and CRC02_patch_embeddings/ populated.
do_generate = True # set True for first run
if do_generate:
loki2.immstain.generate_patch_json_and_embeddings(cfg)
else:
print("Skipping generation; set do_generate=True to run.")
Loading global embeddings...
Global embeddings: 1038158 cells, dim=1280
Loading global nucleus JSON...
../data/imm_stain/CRC02-HE.ome.tif level 0 zarr shape: (3, 54363, 82230)
../data/imm_stain/CRC02-HE.ome.tif level 1 zarr shape: (3, 27182, 41115)
Level0: H0=54363, W0=82230
Level1: H1=27182, W1=41115
Scale factors: scale_y=0.500009, scale_x=0.500000
Patch grid (level1): 14 rows x 21 cols
Assigning global cells to patches (level1 grid)...
Non-empty patches: 188
Saving patch JSON + embedding files...
Done generating patch-specific JSON + embeddings.
Inspect available patches
List patches that have both nuclei/embedding JSON and IF patch present.
available_patches = loki2.immstain.list_available_patches(cfg)
print(f"Found {len(available_patches)} patches with both JSON/emb and IF:")
print(available_patches) # show a sample
# use those patches for quick testing
available_patches=['r004_c006','r004_c005','r004_c007','r003_c006']
Found 4 patches with both JSON/emb and IF:
['r003_c006', 'r004_c005', 'r004_c006', 'r004_c007']
Define train/val/test patch splits
Select a validation patch (held out from train) and optional test patches for final evaluation/visualization.
# Simple deterministic split example (edit as needed)
if len(available_patches) < 3:
raise RuntimeError("Need at least 3 patches to run train/val/test example.")
val_patch = available_patches[0]
train_patches = available_patches[1:-1]
test_patches = [available_patches[-1]]
print("Validation patch:", val_patch)
print("Train patches (count):", len(train_patches))
print("Test patches:", test_patches)
Validation patch: r004_c006
Train patches (count): 2
Test patches: ['r003_c006']
Build datasets
Step 2: Build train/val/test cell-level datasets for a chosen IF channel
Creates cell-level feature matrix X, targets y, and metadata for the chosen IF channel.
with loki2.immstain.timed_section(f"build train dataset (channel {target_channel})"):
X_train, y_train, meta_train = loki2.immstain.build_dataset_for_patches(
cfg,
train_patches,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei
)
with loki2.immstain.timed_section(f"build val dataset (channel {target_channel})"):
X_val, y_val, meta_val = loki2.immstain.build_dataset_for_patches(
cfg,
[val_patch],
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei
)
print("Train shape", X_train.shape, "Val shape", X_val.shape)
Processing patch r004_c005...
N nuclei in patch JSON: 16911
Patch embeddings shape: (16911, 1280)
Processing patch r004_c007...
N nuclei in patch JSON: 16068
Patch embeddings shape: (16068, 1280)
Total cells across all patches: 32979, embedding dim=1280
[timer] build train dataset (channel 2): 122.62s
Processing patch r004_c006...
N nuclei in patch JSON: 15974
Patch embeddings shape: (15974, 1280)
Total cells across all patches: 15974, embedding dim=1280
[timer] build val dataset (channel 2): 49.36s
Train shape (32979, 1280) Val shape (15974, 1280)
Preprocess features and targets
Optionally L2-normalize embeddings and log-transform targets (log1p).
def prep_features(X):
if not normalize_embeddings:
return X.copy()
return loki2.immstain.l2_normalize_embeddings(X.copy())
prep_target = (np.log1p if use_log else (lambda x: x.copy()))
X_train_proc = prep_features(X_train)
X_val_proc = prep_features(X_val)
y_train_proc = prep_target(y_train)
y_val_proc = prep_target(y_val)
print("Preprocessed shapes:", X_train_proc.shape, X_val_proc.shape)
Preprocessed shapes: (32979, 1280) (15974, 1280)
Train LightGBM with patch-level validation
Step 3: Train a LightGBM regressor with per-patch holdout validation
Only the LightGBM model is used to keep the pipeline focused.
with loki2.immstain.timed_section("train lightgbm"):
model, val_metrics, y_val_pred = loki2.immstain.train_lightgbm_regressor(
X_train_proc,
y_train_proc,
X_val_proc,
y_val_proc,
random_state=42,
num_boost_round=2000,
early_stopping_rounds=100,
)
print("Validation metrics:", json.dumps(val_metrics, indent=2))
# Train predictions (for diagnostics on processed scale)
y_train_pred = loki2.immstain.predict_with_model(model, X_train_proc)
Training until validation scores don't improve for 100 rounds
[100] train's rmse: 0.463894 valid's rmse: 0.642342
Early stopping, best iteration is:
[69] train's rmse: 0.496389 valid's rmse: 0.642156
[timer] train lightgbm: 653.69s
Validation metrics: {
"mse": 0.41236442882565294,
"mae": 0.5030152276674281,
"r2": 0.11654008841836616,
"pearson_r": 0.3525812000093537,
"ssim": 0.1934759306385396,
"model_type": "lightgbm"
}
Optional: test set evaluation
Evaluate on held-out patches (if provided) and collect predictions.
test_results = None
if test_patches:
with loki2.immstain.timed_section(f"build test dataset (channel {target_channel})"):
X_test, y_test, meta_test = loki2.immstain.build_dataset_for_patches(
cfg,
test_patches,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei
)
X_test_proc = prep_features(X_test)
y_test_proc = prep_target(y_test)
with loki2.immstain.timed_section("predict test"):
y_test_pred = loki2.immstain.predict_with_model(model, X_test_proc)
if use_log:
y_test_eval = y_test
y_test_pred_eval = np.expm1(y_test_pred)
else:
y_test_eval = y_test
y_test_pred_eval = y_test_pred
test_metrics = loki2.immstain.compute_regression_metrics(y_test_eval, y_test_pred_eval)
print("Test metrics (original scale):", json.dumps(test_metrics, indent=2))
test_results = {
"X": X_test_proc,
"y_true": y_test_proc,
"y_pred": y_test_pred,
"meta": meta_test,
"metrics": test_metrics,
"y_true_raw": y_test_eval,
"y_pred_raw": y_test_pred_eval,
}
else:
print("No test patches provided; skip test evaluation.")
Processing patch r003_c006...
N nuclei in patch JSON: 14871
Patch embeddings shape: (14871, 1280)
Total cells across all patches: 14871, embedding dim=1280
[timer] build test dataset (channel 2): 45.82s
[timer] predict test: 0.03s
Test metrics (original scale): {
"mse": 9694314.333038595,
"mae": 1913.4060172446673,
"r2": 0.13078191878753087,
"pearson_r": 0.4336573916030431,
"ssim": 0.37258682884705424
}
Save model and predictions
Step 4: Save per-cell predictions to JSON and plot diagnostics (pred vs. true, residuals)
Persist the trained model plus per-cell prediction JSON for train/val/test splits.
# Save model bundle
model_out_path.parent.mkdir(parents=True, exist_ok=True)
torch.save(
{
"model": model,
"normalize_embeddings": normalize_embeddings,
"use_log": use_log,
"metrics": val_metrics,
},
model_out_path,
)
print(f"Saved model to {model_out_path}")
# Save train predictions (processed scale)
loki2.immstain.save_prediction_json(
train_pred_path,
channel_idx=target_channel,
extend_nuclei=extend_nuclei,
normalize_embeddings=normalize_embeddings,
metrics=val_metrics,
meta=meta_train,
y_true=y_train_proc,
y_pred=y_train_pred,
)
# Save test predictions if available
loki2.immstain.save_prediction_json(
test_pred_path,
channel_idx=target_channel,
extend_nuclei=extend_nuclei,
normalize_embeddings=normalize_embeddings,
metrics=(test_results or {}).get("metrics", val_metrics),
meta=(test_results or {}).get("meta", []),
y_true=(test_results or {}).get("y_true", []),
y_pred=(test_results or {}).get("y_pred", []),
)
Saved model to ../outputs/imm_stain/models/CRC02_lightgbm_channel2.pt
Per-cell predictions saved to: ../outputs/imm_stain/predictions/CRC02_channel2_train_predictions.json
Per-cell predictions saved to: ../outputs/imm_stain/predictions/CRC02_channel2_test_predictions.json
Visualize predictions
Quick scatter and residual plots on validation predictions (processed scale). For raw-scale plots, invert log1p before plotting.
Patch-level GT vs Pred visualization
Render one patch with per-cell measured IF intensity, predicted intensity, residual heatmap, and scatter. Uses the saved prediction JSON.
patch_for_viz = 'r004_c007'
pred_json_for_viz = train_pred_path
out_path = Path("../outputs/imm_stain") / "figures_viz_only" / f"patch_{patch_for_viz}_ch{target_channel}_tutorial.png"
fig, axes = loki2.immstain.visualize_patch_prediction(
cfg,
patch_name=patch_for_viz,
prediction_json_path=pred_json_for_viz,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei,
predictions_are_log=use_log,
colorbar_range=None,
save_path=out_path
)
plt.show()
print(f"Saved patch viz to {out_path}")
Saved patch visualization to ../outputs/imm_stain/figures_viz_only/patch_r004_c007_ch2_tutorial.png
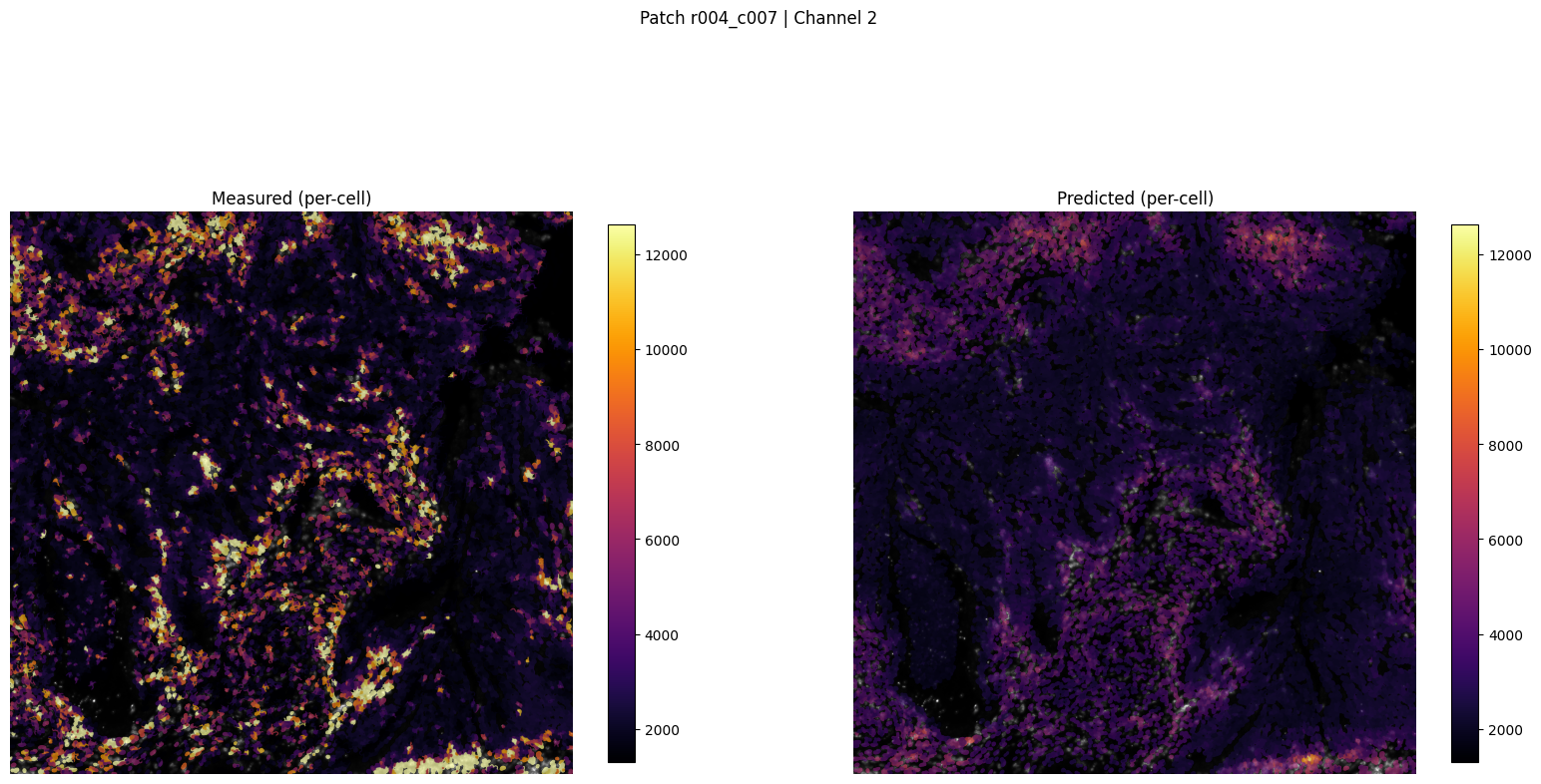
Saved patch viz to ../outputs/imm_stain/figures_viz_only/patch_r004_c007_ch2_tutorial.png
patch_for_viz = test_patches[0] if test_patches else val_patch
pred_json_for_viz = test_pred_path if test_patches else train_pred_path
out_path = Path("../outputs/imm_stain") / "figures_viz_only" / f"patch_{patch_for_viz}_ch{target_channel}_tutorial.png"
fig, axes = loki2.immstain.visualize_patch_prediction(
cfg,
patch_name=patch_for_viz,
prediction_json_path=pred_json_for_viz,
channel_idx=target_channel,
guide_channel=None,
expansion_distance=expansion_distance,
extend_nuclei=extend_nuclei,
predictions_are_log=use_log,
colorbar_range=None,
save_path=out_path
)
plt.show()
print(f"Saved patch viz to {out_path}")
Saved patch visualization to ../outputs/imm_stain/figures_viz_only/patch_r003_c006_ch2_tutorial.png
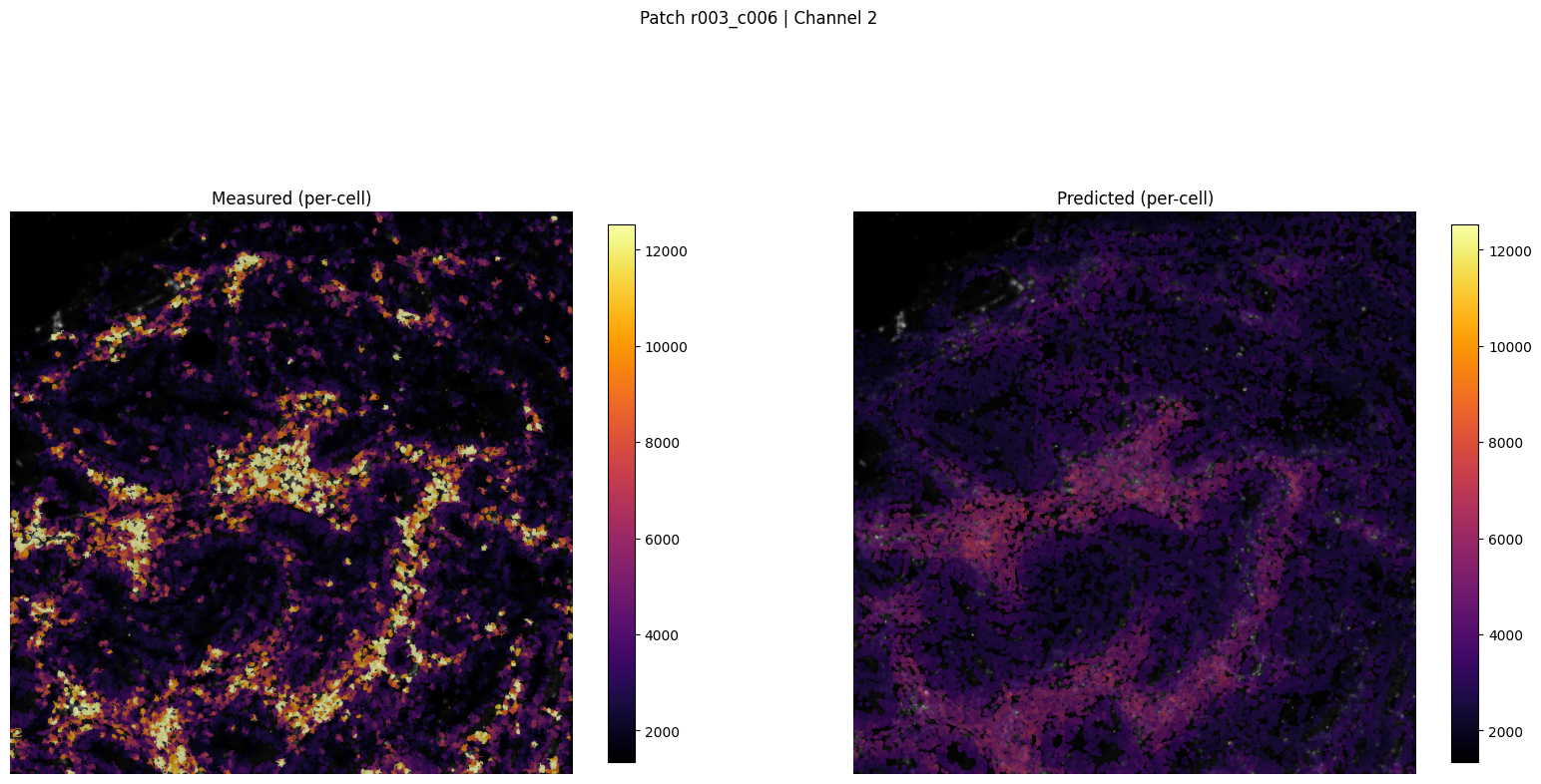
Saved patch viz to ../outputs/imm_stain/figures_viz_only/patch_r003_c006_ch2_tutorial.png