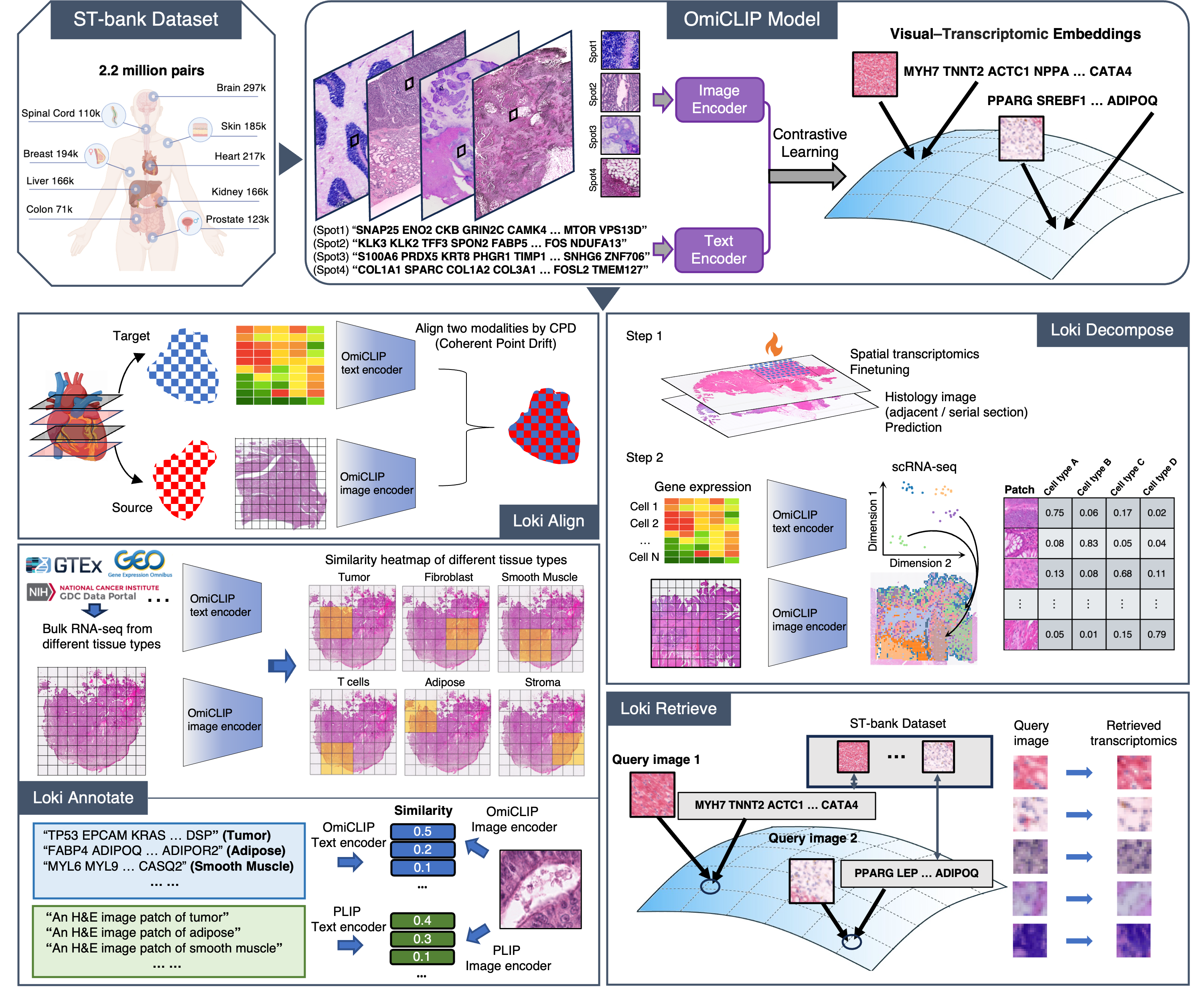
Loki documentation
Building on OmiCLIP, a visual–omics foundation model designed to bridge omics data and hematoxylin and eosin (H&E) images, we developed the Loki platform, which has five key functions: tissue alignment using ST or H&E images, tissue annotation of ST or H&E images based on bulk RNA-seq or marker genes, cell type decomposition of ST or H&E images using scRNA-seq as a reference, histology image–transcriptomics retrieval, and ST gene expression prediction from H&E images.
Please find our Nature Methods paper “A visual–omics foundation model to bridge histopathology with spatial transcriptomics” here.
Please find our github here.
ST-bank database
Pretrained weights
Loki