Basic Usage of OmiCLIP Model
This notebook demonstrates the basic usage of OmiCLIP model, including loading model, encoding text and image, calculating similairty between image embeddings and text embeddings, and examples of preprocess ST data, scRNA-seq data, bulk RNA-seq data, and whole image. It takes about 30 mins to run this notebook using cpu on MacBook Pro.
[1]:
import os
import pandas as pd
import numpy as np
import scanpy as sc
import anndata
from PIL import Image
import loki.utils
import loki.preprocess
/opt/anaconda3/envs/loki_env/lib/python3.9/site-packages/timm/models/layers/__init__.py:48: FutureWarning: Importing from timm.models.layers is deprecated, please import via timm.layers
warnings.warn(f"Importing from {__name__} is deprecated, please import via timm.layers", FutureWarning)
The sample data are stored in the directory data/basic_usage, which can be donwloaded from Google Drive link.
[2]:
data_dir = './data/basic_usage/'
Load OmiCLIP Model
The pretrained weights are avaliable on Hugging Face.
[3]:
model_path = os.path.join(data_dir, 'checkpoint.pt')
device = 'cpu'
[4]:
model, preprocess, tokenizer = loki.utils.load_model(model_path, device)
[5]:
model.eval()
[5]:
CoCa(
(text): TextTransformer(
(token_embedding): Embedding(49408, 768)
(transformer): Transformer(
(resblocks): ModuleList(
(0-11): 12 x ResidualAttentionBlock(
(ln_1): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(attn): MultiheadAttention(
(out_proj): NonDynamicallyQuantizableLinear(in_features=768, out_features=768, bias=True)
)
(ls_1): Identity()
(ln_2): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(mlp): Sequential(
(c_fc): Linear(in_features=768, out_features=3072, bias=True)
(gelu): GELU(approximate='none')
(c_proj): Linear(in_features=3072, out_features=768, bias=True)
)
(ls_2): Identity()
)
)
)
(ln_final): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
)
(visual): VisionTransformer(
(conv1): Conv2d(3, 1024, kernel_size=(14, 14), stride=(14, 14), bias=False)
(patch_dropout): Identity()
(ln_pre): LayerNorm((1024,), eps=1e-05, elementwise_affine=True)
(transformer): Transformer(
(resblocks): ModuleList(
(0-23): 24 x ResidualAttentionBlock(
(ln_1): LayerNorm((1024,), eps=1e-05, elementwise_affine=True)
(attn): MultiheadAttention(
(out_proj): NonDynamicallyQuantizableLinear(in_features=1024, out_features=1024, bias=True)
)
(ls_1): Identity()
(ln_2): LayerNorm((1024,), eps=1e-05, elementwise_affine=True)
(mlp): Sequential(
(c_fc): Linear(in_features=1024, out_features=4096, bias=True)
(gelu): GELU(approximate='none')
(c_proj): Linear(in_features=4096, out_features=1024, bias=True)
)
(ls_2): Identity()
)
)
)
(attn_pool): AttentionalPooler(
(attn): MultiheadAttention(
(out_proj): NonDynamicallyQuantizableLinear(in_features=768, out_features=768, bias=True)
)
(ln_q): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(ln_k): LayerNorm((1024,), eps=1e-05, elementwise_affine=True)
)
(ln_post): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
)
(text_decoder): MultimodalTransformer(
(resblocks): ModuleList(
(0-11): 12 x ResidualAttentionBlock(
(ln_1): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(attn): MultiheadAttention(
(out_proj): NonDynamicallyQuantizableLinear(in_features=768, out_features=768, bias=True)
)
(ls_1): Identity()
(ln_2): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(mlp): Sequential(
(c_fc): Linear(in_features=768, out_features=3072, bias=True)
(gelu): GELU(approximate='none')
(c_proj): Linear(in_features=3072, out_features=768, bias=True)
)
(ls_2): Identity()
)
)
(cross_attn): ModuleList(
(0-11): 12 x ResidualAttentionBlock(
(ln_1): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(attn): MultiheadAttention(
(out_proj): NonDynamicallyQuantizableLinear(in_features=768, out_features=768, bias=True)
)
(ls_1): Identity()
(ln_1_kv): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(ln_2): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
(mlp): Sequential(
(c_fc): Linear(in_features=768, out_features=3072, bias=True)
(gelu): GELU(approximate='none')
(c_proj): Linear(in_features=3072, out_features=768, bias=True)
)
(ls_2): Identity()
)
)
(ln_final): LayerNorm((768,), eps=1e-05, elementwise_affine=True)
)
)
Encode Image
Use OmiCLIP to encode image.
[6]:
image_path = os.path.join(data_dir, 'demo_data', 'TUM-TCGA-TLSHWGSQ.tif')
image = Image.open(image_path)
image
[6]:
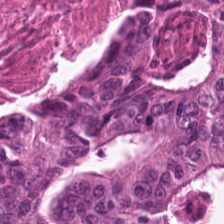
[7]:
image_embeddings = loki.utils.encode_images(model, preprocess, [image_path], device)
image_embeddings.shape
[7]:
torch.Size([1, 768])
Encode Text
Use OmiCLIP to encode text.
[8]:
text = ['TP53 EPCAM KRAS EGFR DEFA5 DEFA6 CEACAM5 CEA KRT18 KRT8 KRT19 CDH17 CK20 MYO6 TP53BP2 PLA2G2A CLDN7 TJP1 PKP3 DSP']
text_embeddings = loki.utils.encode_texts(model, tokenizer, text, device)
text_embeddings.shape
[8]:
torch.Size([1, 768])
Calculate Similarity
Calculate similairty between image embeddings and text embeddings.
[9]:
dot_similarity = image_embeddings @ text_embeddings.T
[10]:
dot_similarity
[10]:
tensor([[0.3926]])
Examples of preprocessing ST data, scRNA-seq data, bulk RNA-seq data, and whole image
Examples of using .h5ad object (ST data or scRNA-seq data) and .gct object (bulk RNA-seq data) to generate text embeddings, and examples of using whole image to generate image patch embeddings.
Encode transcriptome from AnnData object (ST data or scRNA-seq data)
Example of encoding transcriptome from .h5ad object. House keeping gene list is obtained from the Molecular Signatures Database (MSigDB): https://www.gsea-msigdb.org/gsea/msigdb/. It takes about 3 mins to run this section using cpu on MacBook Pro.
[11]:
ad_path = os.path.join(data_dir, 'demo_data', 'RZ_GT_P2.h5ad')
ad = sc.read_h5ad(ad_path)
ad
[11]:
AnnData object with n_obs × n_vars = 3538 × 14467
obs: 'n_counts', 'n_genes', 'percent.mt', 'Adipocyte', 'Cardiomyocyte', 'Endothelial', 'Fibroblast', 'Lymphoid', 'Mast', 'Myeloid', 'Neuronal', 'Pericyte', 'Cycling.cells', 'vSMCs', 'cell_type_original', 'assay_ontology_term_id', 'cell_type_ontology_term_id', 'development_stage_ontology_term_id', 'disease_ontology_term_id', 'ethnicity_ontology_term_id', 'is_primary_data', 'organism_ontology_term_id', 'sex_ontology_term_id', 'tissue_ontology_term_id', 'leiden', 'pixel_y', 'pixel_x', 'cell_type'
var: 'features'
uns: 'X_approximate_distribution', 'X_normalization', 'default_embedding', 'leiden', 'neighbors', 'schema_version', 'spatial', 'title'
obsm: 'X_pca', 'X_spatial', 'X_umap', 'spatial'
obsp: 'connectivities', 'distances'
[12]:
house_keeping_genes = pd.read_csv(os.path.join(data_dir, 'demo_data', 'housekeeping_genes.csv'), index_col = 0)
top_k_genes_str = loki.preprocess.generate_gene_df(ad, house_keeping_genes)
top_k_genes_str
[12]:
| label | |
|---|---|
| RZ_GT_P2_AAACAAGTATCTCCCA-1 | TTN MYH7 MB MALAT1 MTRNR2L12 TPM1 DES CRYAB AP... |
| RZ_GT_P2_AAACAATCTACTAGCA-1 | MYH7 MTRNR2L12 TTN CRYAB MYL2 MB TNNT2 DES TPM... |
| RZ_GT_P2_AAACACCAATAACTGC-1 | TTN MYH7 MTRNR2L12 MB MALAT1 CRYAB TNNI3 DES A... |
| RZ_GT_P2_AAACAGAGCGACTCCT-1 | TTN MYH7 MALAT1 MTRNR2L12 MYL2 MB TPM1 MYL3 CR... |
| RZ_GT_P2_AAACAGCTTTCAGAAG-1 | MB MYH7 TTN CRYAB MTRNR2L12 TNNI3 TPM1 MYL2 DE... |
| ... | ... |
| RZ_GT_P2_TTGTTGTGTGTCAAGA-1 | DES MB TCAP MYH7 TPM1 FN1 CMYA5 TNNI3 COL3A1 S... |
| RZ_GT_P2_TTGTTTCACATCCAGG-1 | TTN MYH7 MTRNR2L12 MALAT1 MB CRYAB DES TNNC1 M... |
| RZ_GT_P2_TTGTTTCATTAGTCTA-1 | TTN MALAT1 MYH7 MTRNR2L12 MB DES CRYAB TNNI3 T... |
| RZ_GT_P2_TTGTTTCCATACAACT-1 | MTRNR2L12 MB MYH7 TTN CRYAB IGKC DES MALAT1 TP... |
| RZ_GT_P2_TTGTTTGTGTAAATTC-1 | TTN MTRNR2L12 MYH7 TNNI3 MALAT1 MB DES CRYAB T... |
3538 rows × 1 columns
[13]:
text_embeddings = loki.utils.encode_text_df(model, tokenizer, top_k_genes_str, 'label', device)
text_embeddings.shape
[13]:
torch.Size([3538, 768])
Encode transcriptome from GCT object (bulk RNA-seq data)
Example of encoding transcriptome from .gct object.
[14]:
sc_data_path = os.path.join(data_dir, 'demo_data', 'bulk_fibroblasts.gct')
gct_data = loki.preprocess.read_gct(sc_data_path)
gct_data
[14]:
| id | Name | Description | GTEX-111VG-0008-SM-5Q5BG | GTEX-111YS-0008-SM-5Q5BH | GTEX-1122O-0008-SM-5QGR2 | GTEX-1128S-0008-SM-5Q5DP | GTEX-113IC-0008-SM-5QGRF | GTEX-113JC-0008-SM-5QGR6 | GTEX-117XS-0008-SM-5Q5DQ | ... | GTEX-ZVE2-0008-SM-51MRU | GTEX-ZVP2-0008-SM-51MSL | GTEX-ZVT2-0008-SM-57WC9 | GTEX-ZVT3-0008-SM-51MRI | GTEX-ZVT4-0008-SM-57WCA | GTEX-ZVTK-0008-SM-57WDA | GTEX-ZVZP-0008-SM-51MSX | GTEX-ZVZQ-0008-SM-51MSK | GTEX-ZXES-0008-SM-57WCX | GTEX-ZXG5-0008-SM-57WDB | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | ENSG00000223972.5 | DDX11L1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | ENSG00000227232.5 | WASH7P | 21 | 31 | 16 | 41 | 57 | 46 | 66 | ... | 24 | 62 | 40 | 42 | 64 | 58 | 62 | 52 | 20 | 32 |
| 2 | 2 | ENSG00000278267.1 | MIR6859-1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | 3 | ENSG00000243485.5 | MIR1302-2HG | 0 | 0 | 0 | 0 | 1 | 0 | 0 | ... | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| 4 | 4 | ENSG00000237613.2 | FAM138A | 0 | 0 | 1 | 0 | 0 | 0 | 0 | ... | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 56195 | 56195 | ENSG00000198695.2 | MT-ND6 | 35691 | 34685 | 50128 | 81334 | 60140 | 81957 | 48581 | ... | 60506 | 94854 | 76311 | 87410 | 63940 | 108716 | 70445 | 78721 | 68057 | 53557 |
| 56196 | 56196 | ENSG00000210194.1 | MT-TE | 8 | 19 | 28 | 47 | 31 | 40 | 25 | ... | 35 | 53 | 23 | 34 | 21 | 52 | 29 | 32 | 19 | 27 |
| 56197 | 56197 | ENSG00000198727.2 | MT-CYB | 161038 | 154424 | 138836 | 246421 | 194695 | 225034 | 214565 | ... | 269451 | 296274 | 335286 | 274560 | 186433 | 312236 | 499192 | 258581 | 241541 | 173705 |
| 56198 | 56198 | ENSG00000210195.2 | MT-TT | 0 | 1 | 1 | 6 | 2 | 0 | 2 | ... | 1 | 1 | 3 | 3 | 6 | 3 | 3 | 1 | 1 | 0 |
| 56199 | 56199 | ENSG00000210196.2 | MT-TP | 3 | 0 | 0 | 3 | 1 | 0 | 3 | ... | 1 | 0 | 1 | 1 | 7 | 4 | 1 | 0 | 1 | 2 |
56200 rows × 507 columns
[15]:
bulk_ad = anndata.AnnData(pd.DataFrame(gct_data.iloc[:, 3:].mean(axis=1)).T)
bulk_ad.var.index = gct_data['Description']
bulk_text_feature = loki.preprocess.generate_gene_df(bulk_ad, house_keeping_genes, todense=False)
bulk_text_feature
/opt/anaconda3/envs/loki_env/lib/python3.9/site-packages/anndata/_core/aligned_df.py:68: ImplicitModificationWarning: Transforming to str index.
warnings.warn("Transforming to str index.", ImplicitModificationWarning)
/opt/anaconda3/envs/loki_env/lib/python3.9/site-packages/anndata/_core/aligned_df.py:68: ImplicitModificationWarning: Transforming to str index.
warnings.warn("Transforming to str index.", ImplicitModificationWarning)
[15]:
| label | |
|---|---|
| 0 | FN1 COL1A1 COL1A2 COL6A3 TGFBI THBS1 COL6A2 CO... |
[16]:
text_embeddings = loki.utils.encode_text_df(model, tokenizer, bulk_text_feature, 'label', device)
text_embeddings.shape
[16]:
torch.Size([1, 768])
Encode patches from a whole image
Example of encoding patches from a whole image. It takes about 28 mins to run this section using cpu on MacBook Pro.
[17]:
coord = pd.read_csv(os.path.join(data_dir, 'demo_data', 'coord.csv'), index_col=0)
coord
[17]:
| pixel_x | pixel_y | |
|---|---|---|
| GSM5924033_AAACAACGAATAGTTC-1 | 1579.906543 | 152.803738 |
| GSM5924033_AAACAAGTATCTCCCA-1 | 478.504673 | 1266.355141 |
| GSM5924033_AAACAATCTACTAGCA-1 | 1234.112151 | 219.626168 |
| GSM5924033_AAACACCAATAACTGC-1 | 1541.121497 | 1467.289721 |
| GSM5924033_AAACAGAGCGACTCCT-1 | 581.308412 | 464.485982 |
| ... | ... | ... |
| GSM5924033_TTGTTTCACATCCAGG-1 | 1246.728973 | 1444.859814 |
| GSM5924033_TTGTTTCATTAGTCTA-1 | 1400.000001 | 1489.252338 |
| GSM5924033_TTGTTTCCATACAACT-1 | 1438.785048 | 1155.140188 |
| GSM5924033_TTGTTTGTATTACACG-1 | 1259.345795 | 1778.971964 |
| GSM5924033_TTGTTTGTGTAAATTC-1 | 1131.775702 | 308.878505 |
4975 rows × 2 columns
[18]:
img = Image.open(os.path.join(data_dir, 'demo_data', 'whole_img.png'))
img
[18]:
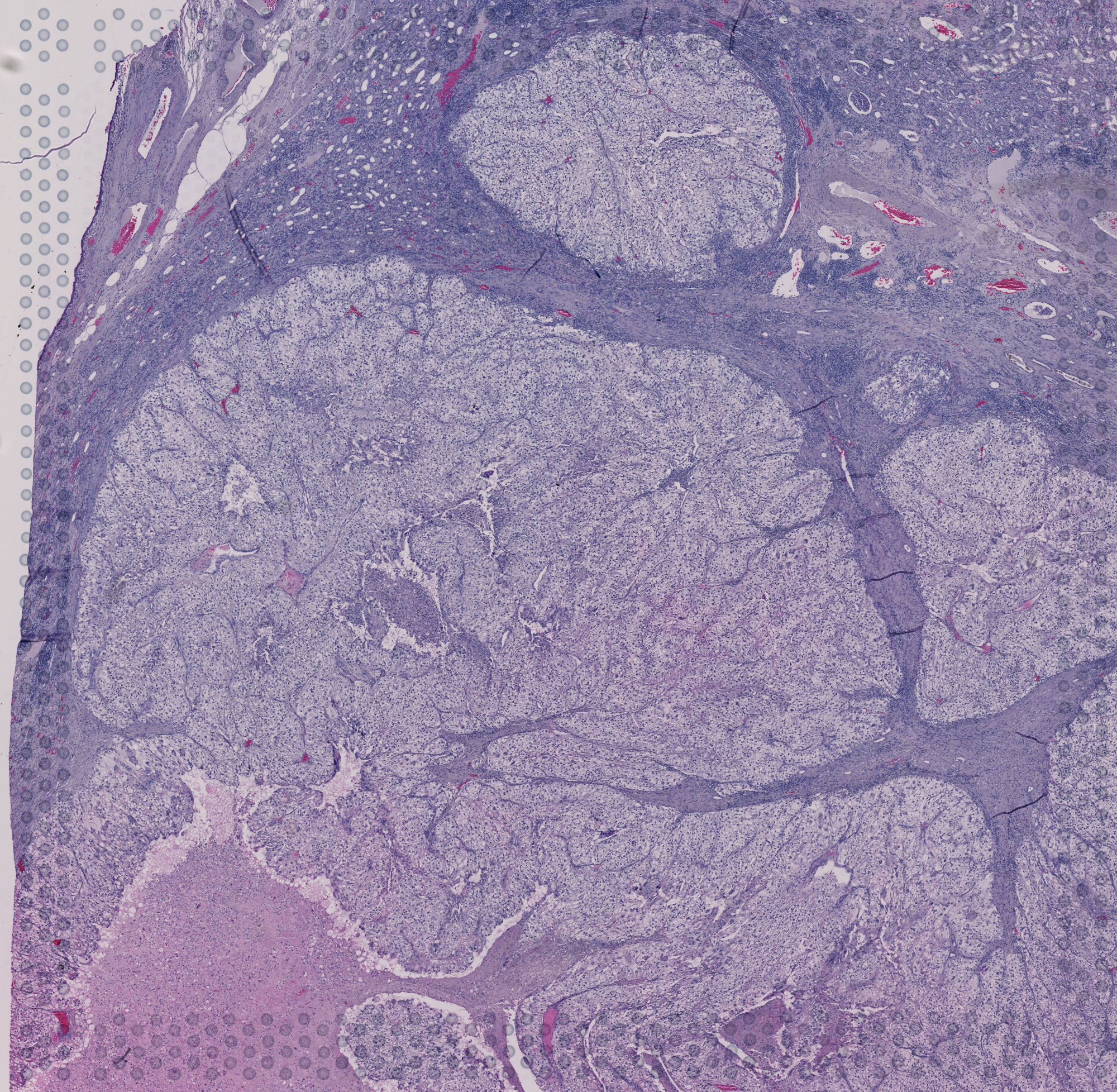
[19]:
img_array = np.asarray(img)
patch_dir = os.path.join(data_dir, 'demo_data', 'patch')
loki.preprocess.segment_patches(img_array, coord, patch_dir)
[20]:
img_list = os.listdir(patch_dir)
patch_paths = [os.path.join(patch_dir, fn) for fn in img_list]
[21]:
image_embeddings = loki.utils.encode_images(model, preprocess, patch_paths, device)
image_embeddings.shape
[21]:
torch.Size([4975, 768])
[ ]: