Loki Decompose - TNBC Sample
This notebook demonstrates how to run Loki Decompose on the in-house triple-negative breast cancer (TNBC) sample. It takes about 1 min to run this notebook on MacBook Pro.
[1]:
import pandas as pd
import os
import scanpy as sc
import loki.decompose
# sc.settings.set_figure_params(dpi=, facecolor="white")
We first finetune OmiCLIP model on the pseudo Visium data in one of four sample region.
[2]:
%%script echo "Comment this line to fine-tune the model on the TNBC data."
import subprocess
import open_clip
model_name='coca_ViT-L-14'
pretrained_weight_path='path to the omiclip pretrained weight'
train_csv = 'visium_data/finetune_data.csv'
name = 'finetune_tnbc'
train_command = [
'python', '-m', 'training.main',
'--name', name,
'--save-frequency', '5',
'--zeroshot-frequency', '10',
'--report-to', 'wandb',
'--train-data', train_csv,
'--csv-img-key', 'img_path',
'--csv-caption-key', 'label',
'--warmup', '10',
'--batch-size', '64',
'--lr', '5e-6',
'--wd', '0.1',
'--epochs', '5',
'--workers', '16',
'--model', model_name,
'--csv-separator', ',',
'--pretrained', pretrained_weight_path,
'--lock-text-freeze-layer-norm',
'--lock-image-freeze-bn-stats',
'--coca-caption-loss-weight','0',
'--coca-contrastive-loss-weight','1',
'--val-frequency', '10',
'--aug-cfg', 'color_jitter=(0.32, 0.32, 0.32, 0.08)', 'color_jitter_prob=0.5', 'gray_scale_prob=0'
]
subprocess.run(train_command)
Comment this line to fine-tune the model on the TNBC data.
We provide the embeddings generated from the OmiCLIP model. The sample data and embeddings are stored in the directory data/loki_decompose/TNBC_data, which can be donwloaded from Google Drive link.
Here is a list of the files that are needed to run the cell type decomposition on the pseudo Visium data:
.
├── checkpoint_tnbc
│ ├── TNBC_img_features_finetune.csv
│ ├── TNBC_txt_features_finetune.csv
│ └── TNBC_txt_features_sc_finetune.csv
├── scRNA_data
│ └── scRNA_data.h5ad
└── pseudo_visium_data
├── TNBC_pseudo_Visium.h5ad
└── TNBC_Xenium_celltype_spot.h5ad
[5]:
data_path = './data/loki_decompose/TNBC_data/'
sample_name = 'TNBC'
[6]:
def generate_deconv_df(sc_ad, st_ad):
for cell_type in st_ad.obsm['tangram_ct_pred'].columns:
st_ad.obsm['tangram_ct_pred'][cell_type]=st_ad.obsm['tangram_ct_pred'][cell_type]*sc_ad.obs['cell_type'].value_counts()[cell_type]
st_ad.obsm['tangram_ct_pred']['Immune']=st_ad.obsm['tangram_ct_pred']['Macrophage']+ \
st_ad.obsm['tangram_ct_pred']['B cell'] + \
st_ad.obsm['tangram_ct_pred']['T cell']
st_ad.obsm['tangram_ct_pred'].drop(['Macrophage','B cell','T cell'],axis=1, inplace=True)
st_ad.obsm['tangram_ct_pred'] = st_ad.obsm['tangram_ct_pred'][['Epithelial', 'Immune', 'Stroma']]
deconv_df = st_ad.obsm['tangram_ct_pred'].T/st_ad.obsm['tangram_ct_pred'].T.sum()
deconv_df = deconv_df.T
return deconv_df
[7]:
sc_data_raw = sc.read_h5ad(os.path.join(data_path, 'scRNA_data', 'TNBC_SC_preprocess.h5ad'))
ad_vis = sc.read_h5ad(os.path.join(data_path, 'pseudo_visium_data', 'TNBC_pseudo_Visium.h5ad'))
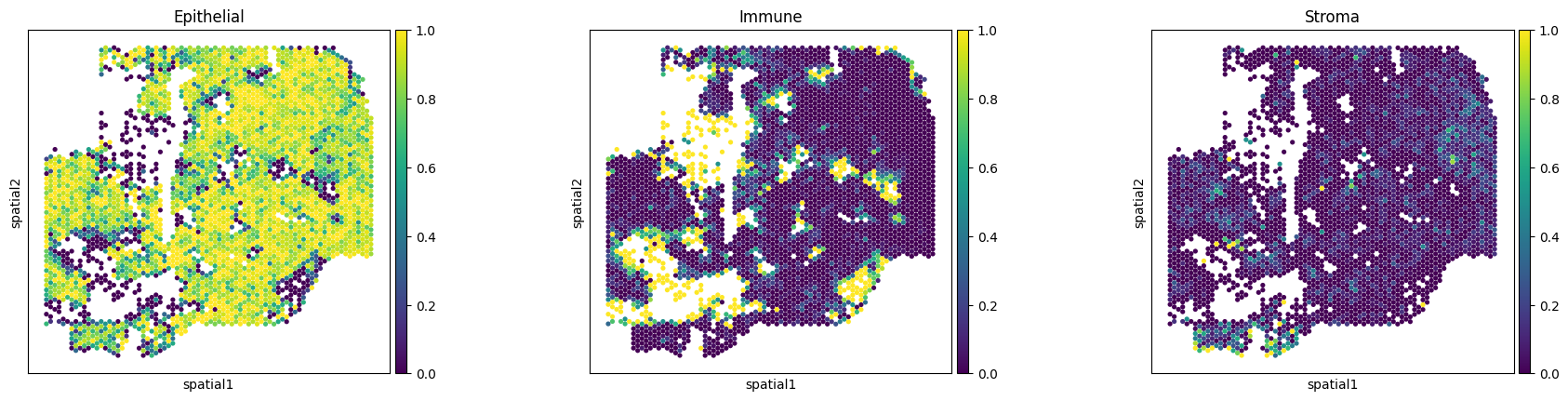
Visualize cell type component defined by Xenium data (ground truth data).
[8]:
ad_xen_sp = sc.read_h5ad(os.path.join(data_path, 'pseudo_visium_data', 'TNBC_Xenium_celltype_spot.h5ad'))
raw_deconv = ad_xen_sp.obs.pivot_table(index='spot', columns='cell_type', aggfunc='size').reindex(ad_vis.obs.index).T
raw_deconv = raw_deconv/raw_deconv.sum(axis=0)
raw_deconv = raw_deconv.T
raw_deconv
[8]:
| cell_type | Epithelial | Immune | Stroma |
|---|---|---|---|
| TNBC_processed_adata_AACACGTGCATCGCAC-1 | 0.000000 | 1.000000 | 0.000000 |
| TNBC_processed_adata_AACACTTGGCAAGGAA-1 | 0.750000 | 0.250000 | 0.000000 |
| TNBC_processed_adata_AACAGGAAGAGCATAG-1 | 0.772727 | 0.000000 | 0.227273 |
| TNBC_processed_adata_AACAGGATTCATAGTT-1 | 0.916667 | 0.083333 | 0.000000 |
| TNBC_processed_adata_AACAGGTTATTGCACC-1 | 0.846154 | 0.153846 | 0.000000 |
| ... | ... | ... | ... |
| TNBC_processed_adata_TGTTGGAACCTTCCGC-1 | 1.000000 | 0.000000 | 0.000000 |
| TNBC_processed_adata_TGTTGGAACGAGGTCA-1 | 0.968750 | 0.000000 | 0.031250 |
| TNBC_processed_adata_TGTTGGAAGCTCGGTA-1 | 0.793103 | 0.000000 | 0.206897 |
| TNBC_processed_adata_TGTTGGATGGACTTCT-1 | 1.000000 | 0.000000 | 0.000000 |
| TNBC_processed_adata_TGTTGGCCTACACGTG-1 | 0.800000 | 0.200000 | 0.000000 |
3402 rows × 3 columns
[12]:
sc.pl.spatial(ad_vis, title='H&E Image')
[13]:
ad_vis_copy = ad_vis.copy()
ad_vis_copy.obs = pd.concat([ad_vis_copy.obs,raw_deconv],axis=1)
[14]:
sc.pl.spatial(ad_vis_copy, color=['Epithelial', 'Immune', 'Stroma'], alpha_img=0, spot_size=400)
Loki Decompose with fintuning
Run cell type decomposition using the pseudo Visium data.
[15]:
case_name = '_finetune'
ad_expr = ad_vis.copy()
sc_data = sc_data_raw.copy()
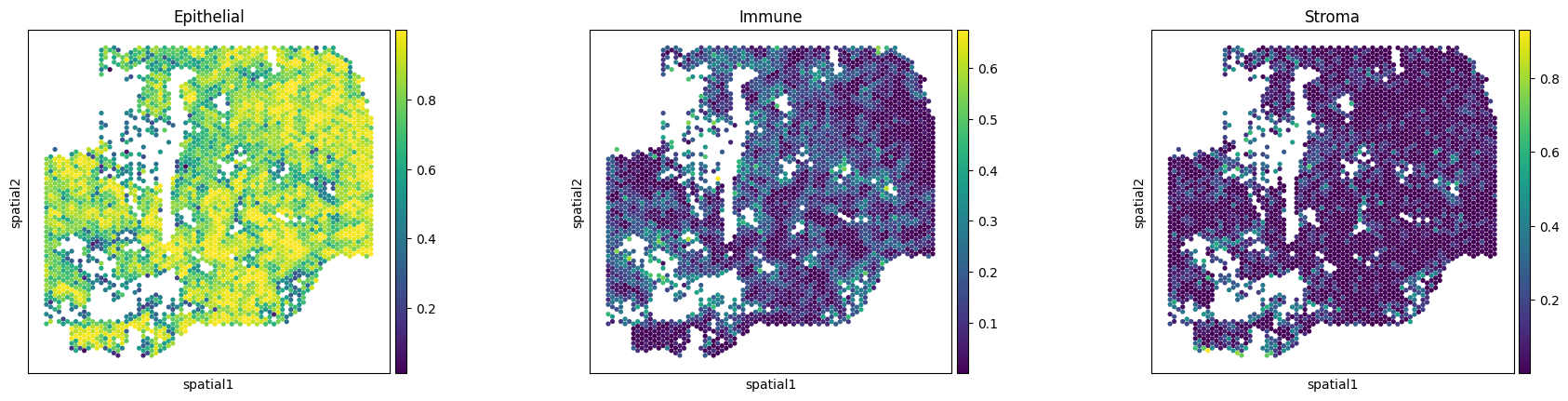
Decompose ST data
Run cell type decomposition using the ST data.
[16]:
sc_text_feature_path = os.path.join(data_path, 'checkpoint_tnbc', sample_name+'_txt_features_sc'+case_name+'.csv')
text_feature_path = os.path.join(data_path, 'checkpoint_tnbc', sample_name+'_txt_features'+case_name+'.csv')
sc_ad = loki.decompose.generate_feature_ad(sc_data, sc_text_feature_path, sc=True)
txt_ad = loki.decompose.generate_feature_ad(ad_expr, text_feature_path)
[17]:
%%capture
txt_ad = loki.decompose.cell_type_decompose(sc_ad, txt_ad)
[18]:
deconv_df = generate_deconv_df(sc_ad, txt_ad)
txt_ad.obs = pd.concat([txt_ad.obs, deconv_df], axis=1)
[19]:
sc.pl.spatial(txt_ad, color=['Epithelial','Immune','Stroma'], alpha_img=0, spot_size=400)
Decompose image
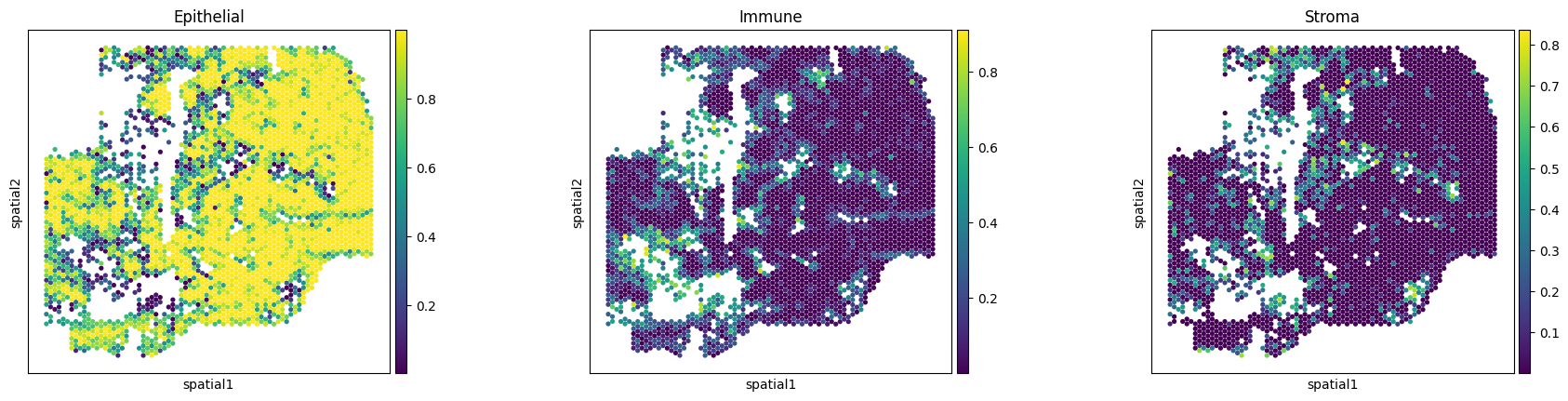
Run cell type decomposition using the image.
[20]:
image_feature_path = os.path.join(data_path, 'checkpoint_tnbc', sample_name+'_img_features'+case_name+'.csv')
sc_ad = loki.decompose.generate_feature_ad(sc_data, sc_text_feature_path, sc=True)
img_ad = loki.decompose.generate_feature_ad(ad_expr, image_feature_path)
[21]:
%%capture
img_ad = loki.decompose.cell_type_decompose(sc_ad, img_ad)
[22]:
deconv_df = generate_deconv_df(sc_ad, img_ad)
img_ad.obs = pd.concat([img_ad.obs, deconv_df], axis=1)
[23]:
sc.pl.spatial(img_ad, color=['Epithelial','Immune','Stroma'], alpha_img=0, spot_size=400)
[ ]: