Loki Align - 8 normal small intestine samples
This notebook demonstrates how to run Loki Align on a dataset of 8 adjunction sections of normal small intestine samples. It takes about 3 mins to run the code to align (ST-to-ST and Image-to-ST) each sample (about 20 mins to finish all 7 source samples) on MacBook Pro .
[1]:
import pandas as pd
import matplotlib.pyplot as plt
import os
from matplotlib.colors import rgb2hex
import loki.align
import loki.preprocess
import loki.utils
import loki.plot
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/timm/models/layers/__init__.py:48: FutureWarning: Importing from timm.models.layers is deprecated, please import via timm.layers
warnings.warn(f"Importing from {__name__} is deprecated, please import via timm.layers", FutureWarning)
We first finetune OmiCLIP model on the target small intestin sample data.
[2]:
%%script echo "Comment this line to fine-tune the model on the target small intestine data."
import subprocess
import open_clip
model_name='coca_ViT-L-14'
pretrained_weight_path='path to the omiclip pretrained weight'
train_csv = 'visium_data/finetune_data.csv'
name = 'finetune_si'
train_command = [
'python', '-m', 'training.main',
'--name', name,
'--save-frequency', '5',
'--zeroshot-frequency', '10',
'--report-to', 'wandb',
'--train-data', train_csv,
'--csv-img-key', 'img_path',
'--csv-caption-key', 'label',
'--warmup', '10',
'--batch-size', '64',
'--lr', '5e-6',
'--wd', '0.1',
'--epochs', '10',
'--workers', '16',
'--model', model_name,
'--csv-separator', ',',
'--pretrained', pretrained_weight_path,
'--lock-text-freeze-layer-norm',
'--lock-image-freeze-bn-stats',
'--coca-caption-loss-weight','0',
'--coca-contrastive-loss-weight','1',
'--val-frequency', '10',
'--aug-cfg', 'color_jitter=(0.32, 0.32, 0.32, 0.08)', 'color_jitter_prob=0.5', 'gray_scale_prob=0'
]
subprocess.run(train_command)
Comment this line to fine-tune the model on the target small intestine data.
We provide the embeddings generated from the OmiCLIP model. The sample data and embeddings are stored in the directory data/loki_align/eight_small_intestine_samples, which can be donwloaded from Google Drive link.
Here is a list of the files that are needed to run the cell type decomposition on the pseudo Visium data:
.
├── checkpoint_si
│ ├── V10F24-048_A1_img_features.csv
│ ├── V10F24-048_A1_txt_features.csv
│ ├── V10F24-048_B1_img_features.csv
│ ├── V10F24-048_B1_txt_features.csv
│ ├── V10F24-048_C1_txt_features.csv
│ ├── V10F24-048_C1_img_features.csv
│ ├── V10F24-048_D1_txt_features.csv
│ ├── V10F24-048_D1_img_features.csv
│ ├── V10F24-050_A1_txt_features_zs.csv
│ ├── V10F24-050_A1_img_features.csv
│ ├── V10F24-050_A1_txt_features.csv
│ ├── V10F24-050_B1_img_features.csv
│ ├── V10F24-050_B1_txt_features.csv
│ ├── V10F24-050_C1_txt_features.csv
│ ├── V10F24-050_C1_img_features.csv
│ ├── V10F24-050_D1_txt_features.csv
│ └── V10F24-050_D1_img_features.csv
└── visium_data
├── V10F24-048_A1.h5ad
├── V10F24-048_B1.h5ad
├── V10F24-048_C1.h5ad
├── V10F24-048_D1.h5ad
├── V10F24-050_A1.h5ad
├── V10F24-050_B1.h5ad
├── V10F24-050_C1.h5ad
└── V10F24-050_D1.h5ad
[3]:
data_dir = './data/loki_align/eight_small_intestine_samples/'
[4]:
def get_features(sample_name, feature_name):
features_path=os.path.join(data_dir, 'checkpoint_si', sample_name+feature_name+'.csv')
features = pd.read_csv(features_path, index_col=0, header = None)
return features
[5]:
tar_sample_name = 'V10F24-050_A1'
ad_path = os.path.join(data_dir, 'visium_data', tar_sample_name+'.h5ad')
ad_tar, ad_tar_coor, tar_img = loki.preprocess.prepare_data_for_alignment(ad_path)
Future exception was never retrieved
future: <Future finished exception=BrokenPipeError(32, 'Broken pipe')>
Traceback (most recent call last):
File "/opt/anaconda3/envs/loki/lib/python3.9/asyncio/unix_events.py", line 665, in write
n = os.write(self._fileno, data)
BrokenPipeError: [Errno 32] Broken pipe
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
Loki Align on ST and image alignment
Run tissue alignment using the ST data and image.
Source1
[6]:
src_sample_name = 'V10F24-050_B1'
ad_path = os.path.join(data_dir, 'visium_data', src_sample_name+'.h5ad')
ad_src, ad_src_coor, src_img = loki.preprocess.prepare_data_for_alignment(ad_path)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
Loki Align on ST to ST alignment
Use Loki Align to align ST to ST.
[7]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[8]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
pca_rgb_comb = (pca_comb_features-pca_comb_features.min(axis=0))/(pca_comb_features.max(axis=0)-pca_comb_features.min(axis=0))
pca_rgb_comb[:,[0,1,2]]=pca_rgb_comb[:,[0,2,1]]
pca_hex_comb = [ rgb2hex(pca_rgb_comb[i,:]) for i in range(pca_rgb_comb.shape[0]) ]
[9]:
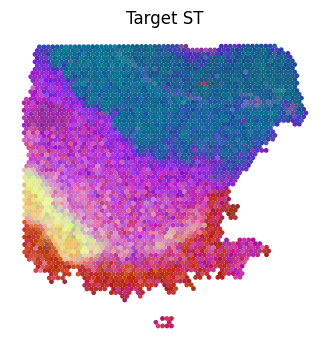
plt.figure(figsize=(4,4))
plt.scatter(ad_tar_coor[:,0], ad_tar_coor[:,1], marker='o', s=6, c=pca_hex_comb[:len(tar_features.T)])
plt.title('Target ST')
plt.axis('off')
plt.show()
[10]:
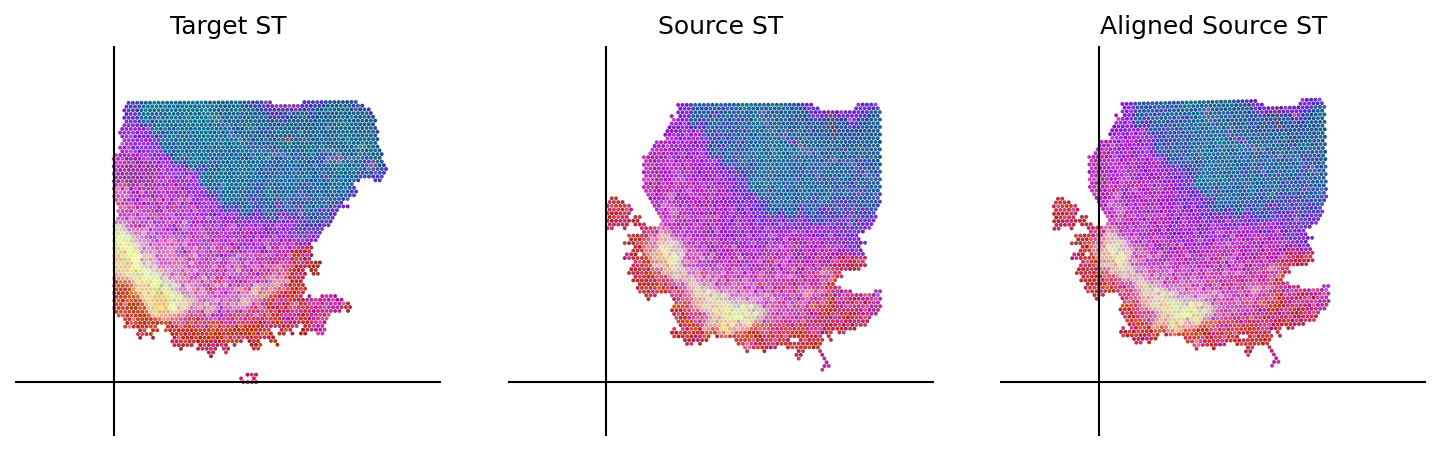
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[11]:
len(pca_hex_comb)
[11]:
6760
[12]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[13]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
[14]:
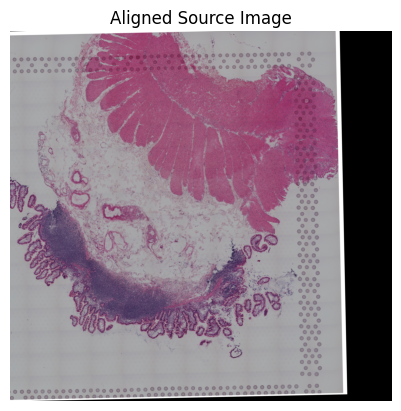
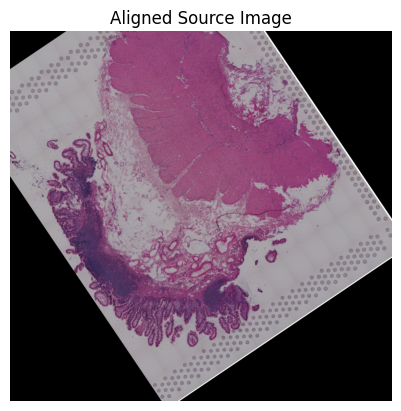
loki.plot.show_image(aligned_image)
Loki Align on image to ST Alignment
Use Loki Align to align image to ST.
[15]:
tar_features = get_features(tar_sample_name, '_txt_features')
src_features = get_features(src_sample_name, '_img_features')
[16]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
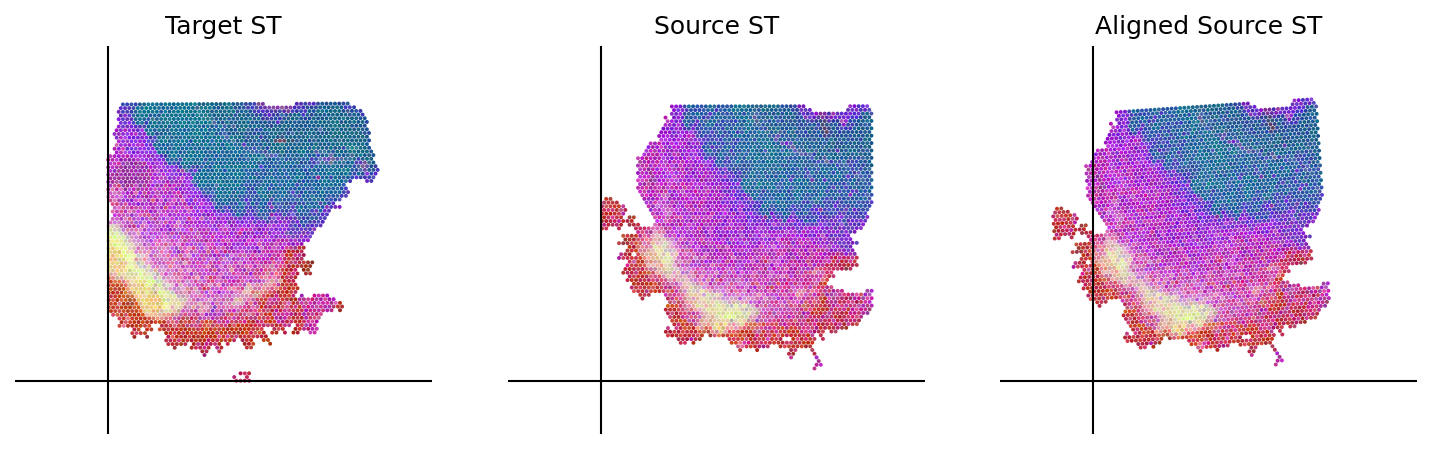
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[17]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
[18]:
loki.plot.show_image(aligned_image)
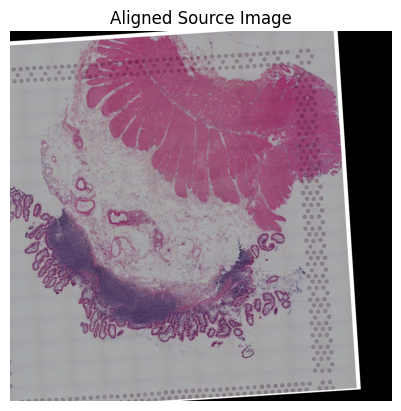
Source2
[19]:
src_sample_name = 'V10F24-050_C1'
ad_path = os.path.join(data_dir, 'visium_data', src_sample_name+'.h5ad')
ad_src, ad_src_coor, src_img = loki.preprocess.prepare_data_for_alignment(ad_path)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
Loki Align on ST to ST alignment
Use Loki Align to align ST to ST.
[20]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[21]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
pca_rgb_comb = (pca_comb_features-pca_comb_features.min(axis=0))/(pca_comb_features.max(axis=0)-pca_comb_features.min(axis=0))
pca_rgb_comb[:,[0,1,2]]=pca_rgb_comb[:,[0,2,1]]
pca_hex_comb = [ rgb2hex(pca_rgb_comb[i,:]) for i in range(pca_rgb_comb.shape[0]) ]
[22]:
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[23]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[24]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
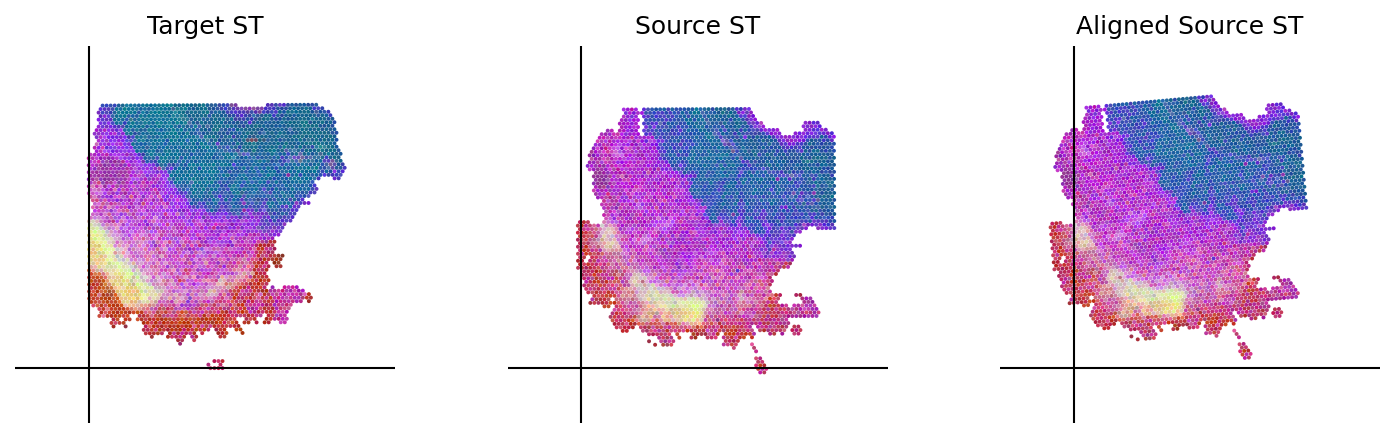
[25]:
loki.plot.show_image(aligned_image)
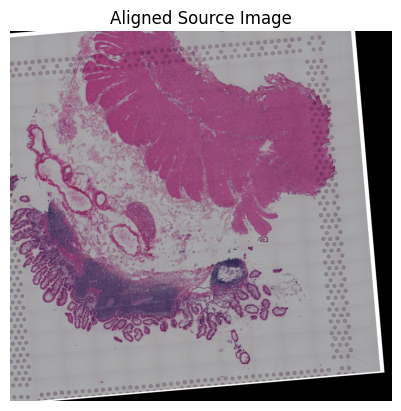
Loki Align on image to ST Alignment
Use Loki Align to align image to ST.
[26]:
tar_features = get_features(tar_sample_name, '_txt_features')
src_features = get_features(src_sample_name, '_img_features')
[27]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[28]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[29]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features, boundary_line=False)
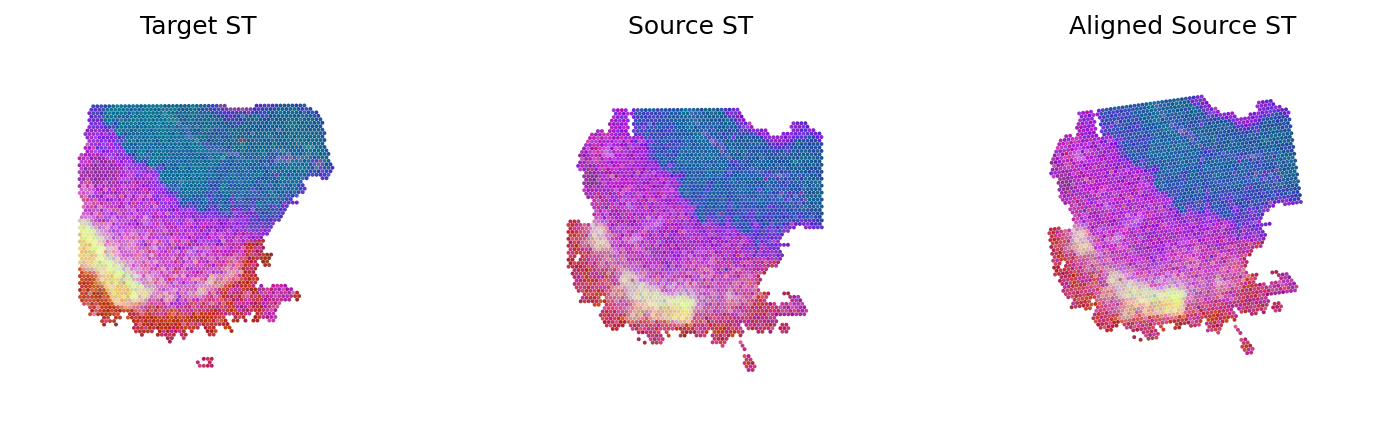
[30]:
loki.plot.show_image(aligned_image)
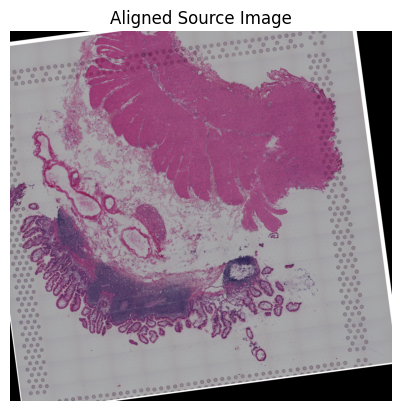
Source3
[31]:
src_sample_name = 'V10F24-050_D1'
ad_path = os.path.join(data_dir, 'visium_data', src_sample_name+'.h5ad')
ad_src, ad_src_coor, src_img = loki.preprocess.prepare_data_for_alignment(ad_path)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
Loki Align on ST to ST alignment
Use Loki Align to align ST to ST.
[32]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[33]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
pca_rgb_comb = (pca_comb_features-pca_comb_features.min(axis=0))/(pca_comb_features.max(axis=0)-pca_comb_features.min(axis=0))
pca_rgb_comb[:,[0,1,2]]=pca_rgb_comb[:,[0,2,1]]
pca_hex_comb = [ rgb2hex(pca_rgb_comb[i,:]) for i in range(pca_rgb_comb.shape[0]) ]
[34]:
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[35]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[36]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
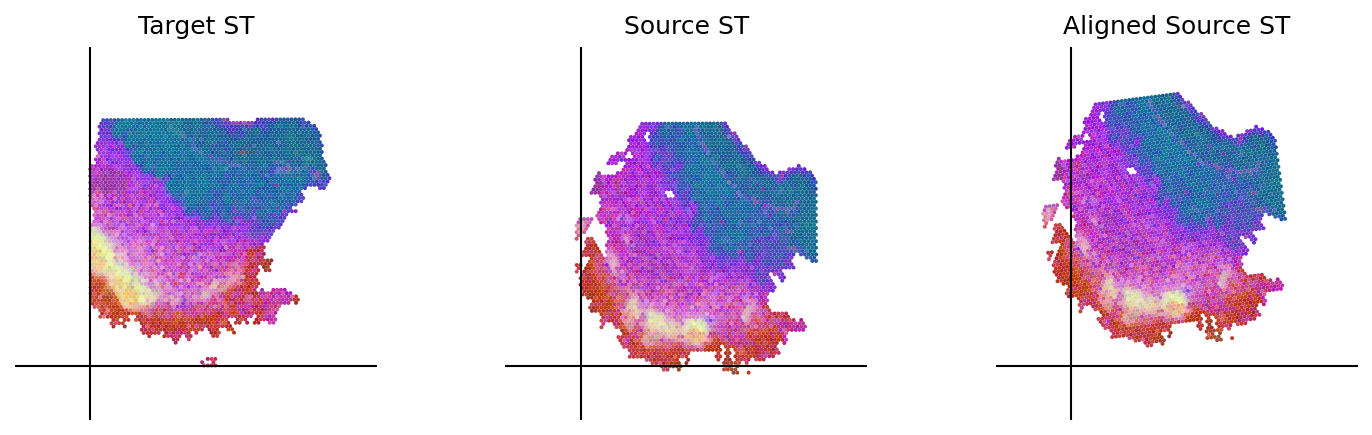
[37]:
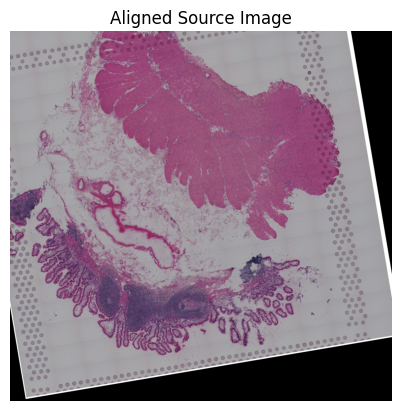
loki.plot.show_image(aligned_image)
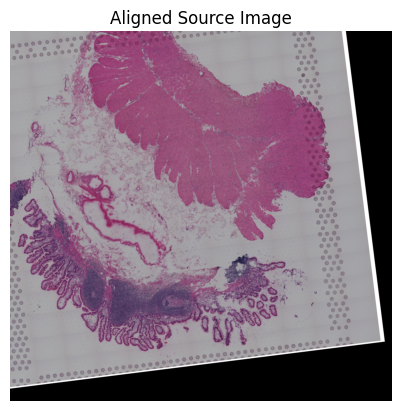
Loki Align on image to ST Alignment
Use Loki Align to align image to ST.
[38]:
tar_features = get_features(tar_sample_name, '_txt_features')
src_features = get_features(src_sample_name, '_img_features')
[39]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[40]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[41]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
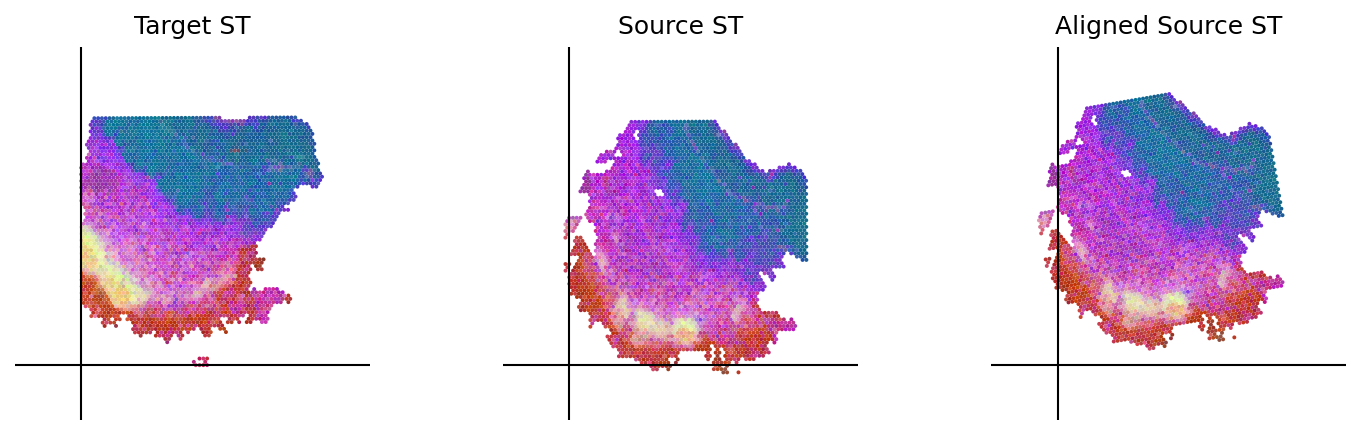
[42]:
loki.plot.show_image(aligned_image)
Source4
[43]:
src_sample_name = 'V10F24-048_C1'
ad_path = os.path.join(data_dir, 'visium_data', src_sample_name+'.h5ad')
ad_src, ad_src_coor, src_img = loki.preprocess.prepare_data_for_alignment(ad_path)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
Loki Align on ST to ST Alignment
Use Loki Align to align ST to ST.
[44]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[45]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
pca_rgb_comb = (pca_comb_features-pca_comb_features.min(axis=0))/(pca_comb_features.max(axis=0)-pca_comb_features.min(axis=0))
pca_rgb_comb[:,[0,1,2]]=pca_rgb_comb[:,[0,2,1]]
pca_hex_comb = [ rgb2hex(pca_rgb_comb[i,:]) for i in range(pca_rgb_comb.shape[0]) ]
[46]:
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[47]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[48]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
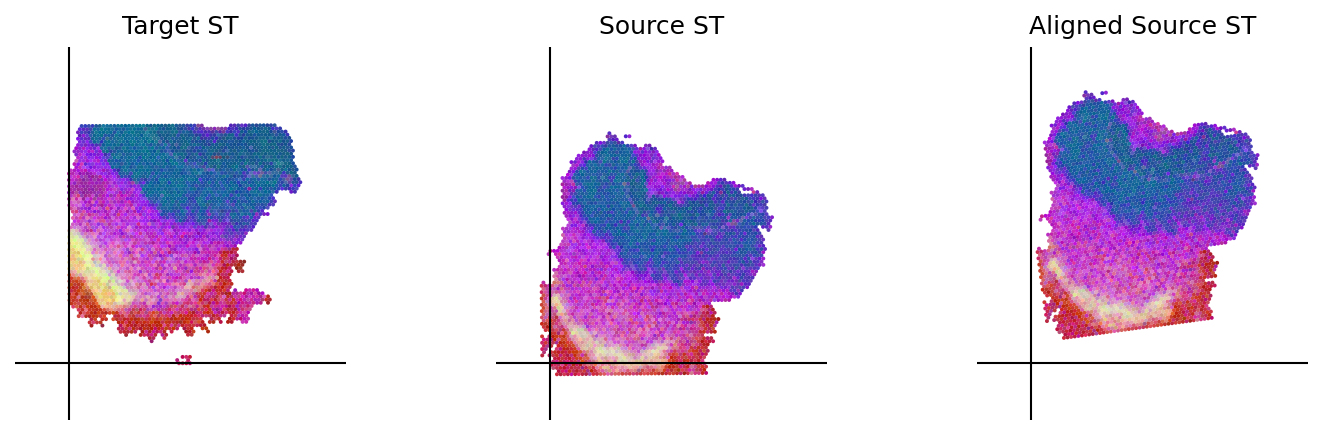
[49]:
loki.plot.show_image(aligned_image)
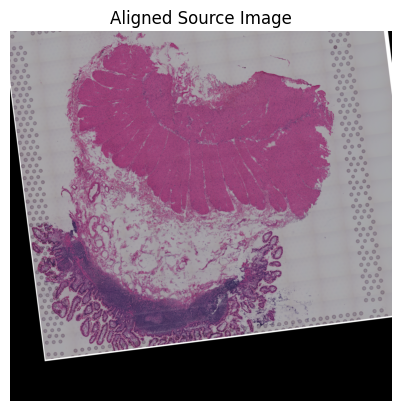
Loki Align on image to ST Alignment
Use Loki Align to align image to ST.
[50]:
tar_features = get_features(tar_sample_name, '_txt_features')
src_features = get_features(src_sample_name, '_img_features')
[51]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[52]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[53]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
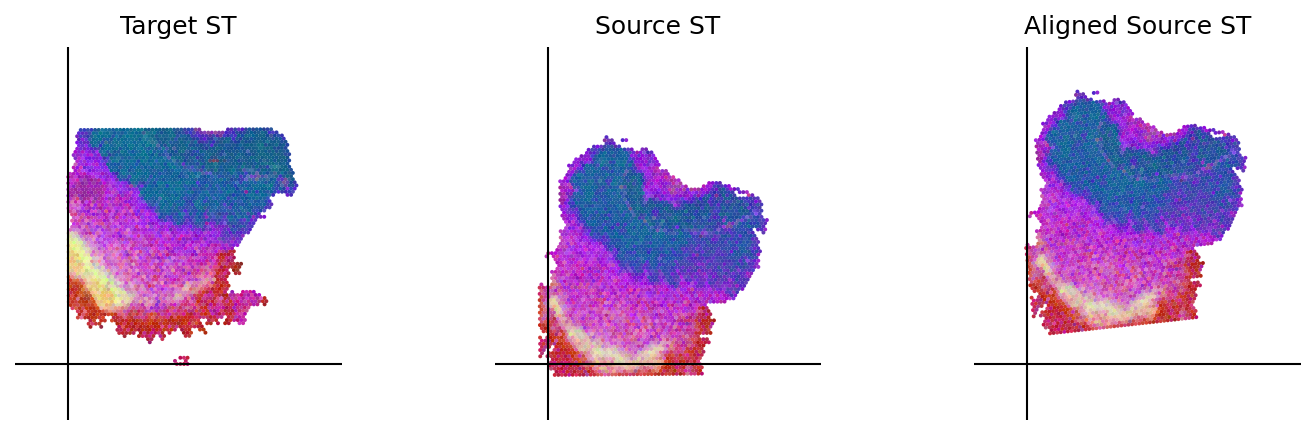
[54]:
loki.plot.show_image(aligned_image)
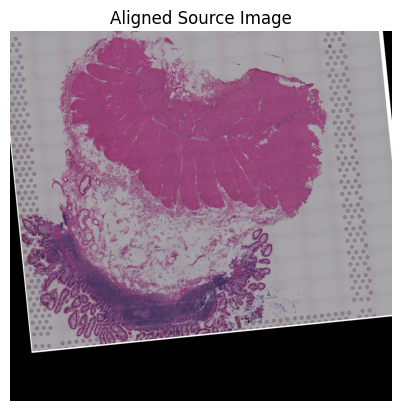
Source5
[55]:
src_sample_name = 'V10F24-048_D1'
ad_path = os.path.join(data_dir, 'visium_data', src_sample_name+'.h5ad')
ad_src, ad_src_coor, src_img = loki.preprocess.prepare_data_for_alignment(ad_path)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
Loki Align on ST to ST alignment
Use Loki Align to align ST to ST.
[56]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[57]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
pca_rgb_comb = (pca_comb_features-pca_comb_features.min(axis=0))/(pca_comb_features.max(axis=0)-pca_comb_features.min(axis=0))
pca_rgb_comb[:,[0,1,2]]=pca_rgb_comb[:,[0,2,1]]
pca_hex_comb = [ rgb2hex(pca_rgb_comb[i,:]) for i in range(pca_rgb_comb.shape[0]) ]
[58]:
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[59]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
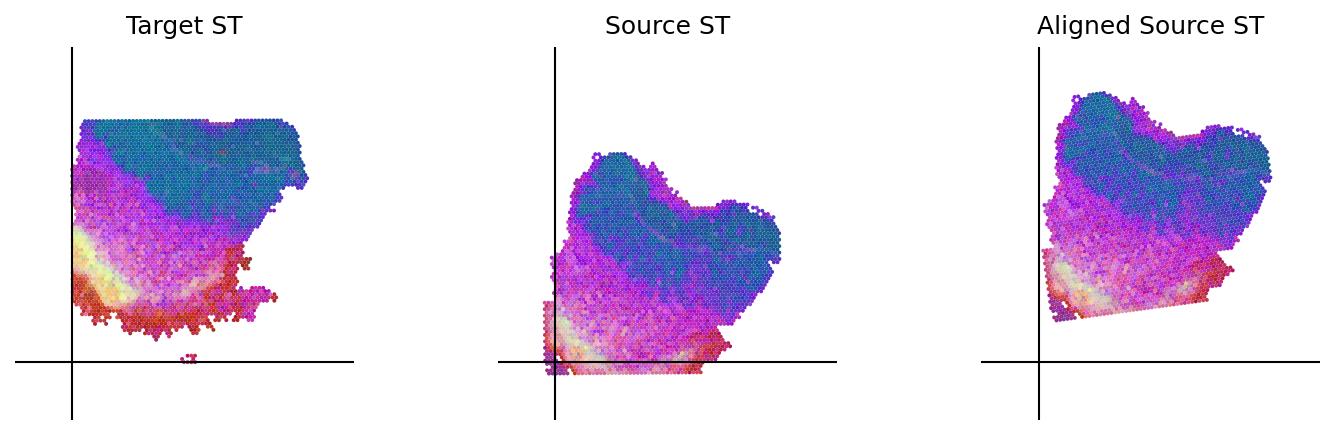
[60]:
loki.plot.show_image(aligned_image)
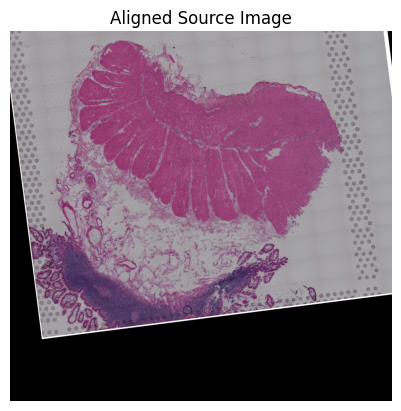
Loki Align on image to ST Alignment
Use Loki Align to align image to ST.
[61]:
tar_features = get_features(tar_sample_name, '_txt_features')
src_features = get_features(src_sample_name, '_img_features')
[62]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[63]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[64]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
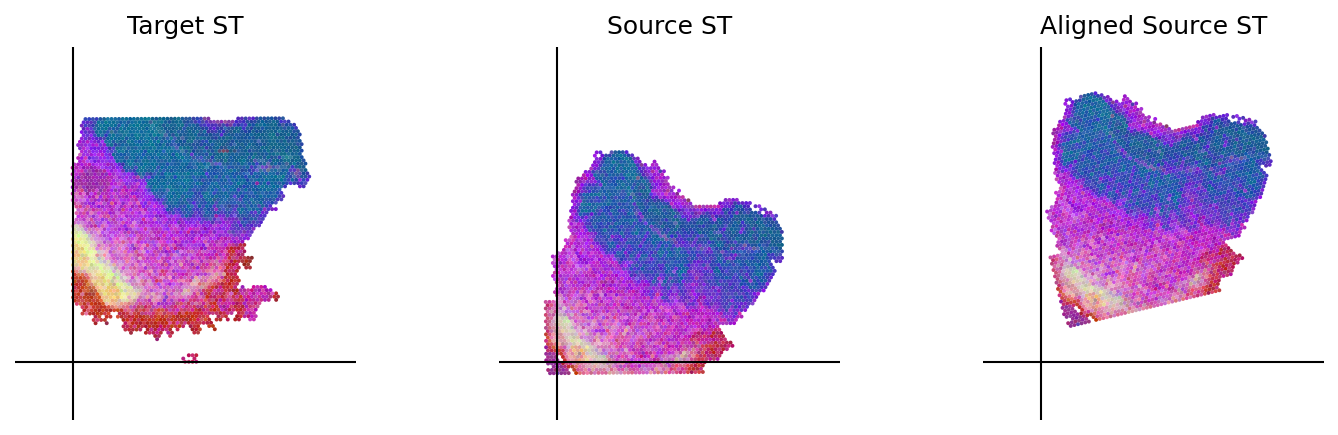
[65]:
loki.plot.show_image(aligned_image)
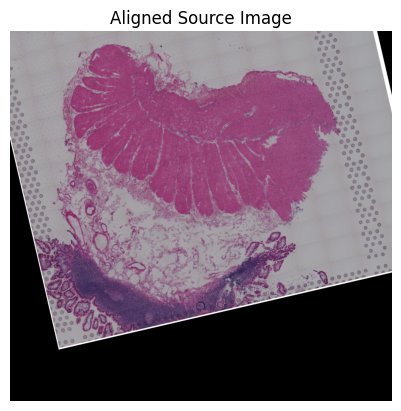
Source6
[66]:
src_sample_name = 'V10F24-048_B1'
ad_path = os.path.join(data_dir, 'visium_data', src_sample_name+'.h5ad')
ad_src, ad_src_coor, src_img = loki.preprocess.prepare_data_for_alignment(ad_path)
Loki Align on ST to ST alignment
Use Loki Align to align ST to ST.
[67]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[68]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
pca_rgb_comb = (pca_comb_features-pca_comb_features.min(axis=0))/(pca_comb_features.max(axis=0)-pca_comb_features.min(axis=0))
pca_rgb_comb[:,[0,1,2]]=pca_rgb_comb[:,[0,2,1]]
pca_hex_comb = [ rgb2hex(pca_rgb_comb[i,:]) for i in range(pca_rgb_comb.shape[0]) ]
[69]:
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[70]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[71]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
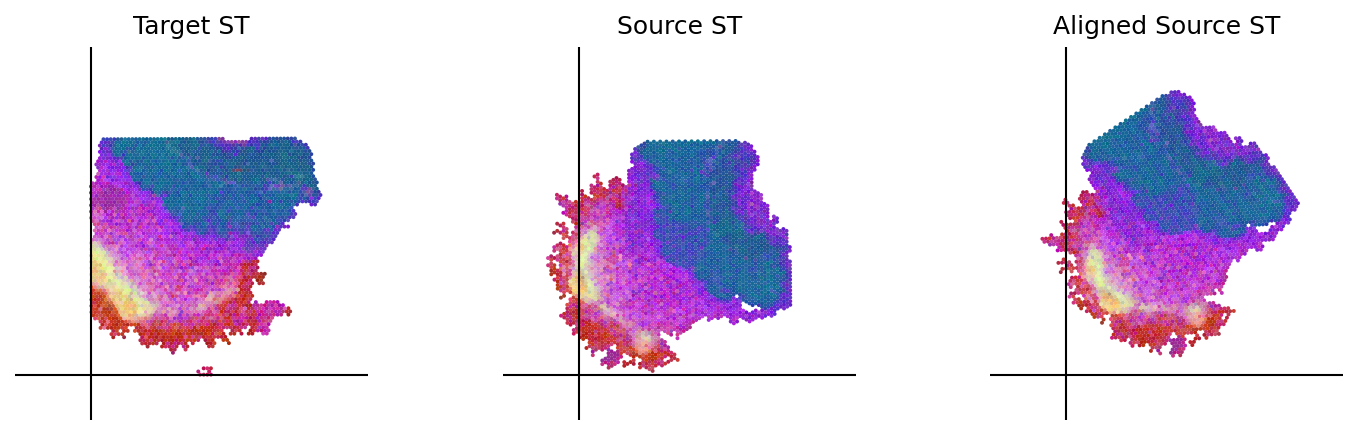
[72]:
loki.plot.show_image(aligned_image)
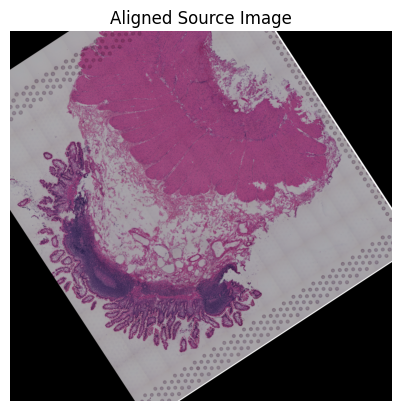
Loki Align on image to ST Alignment
Use Loki Align to align image to ST.
[73]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[74]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[75]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[76]:
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
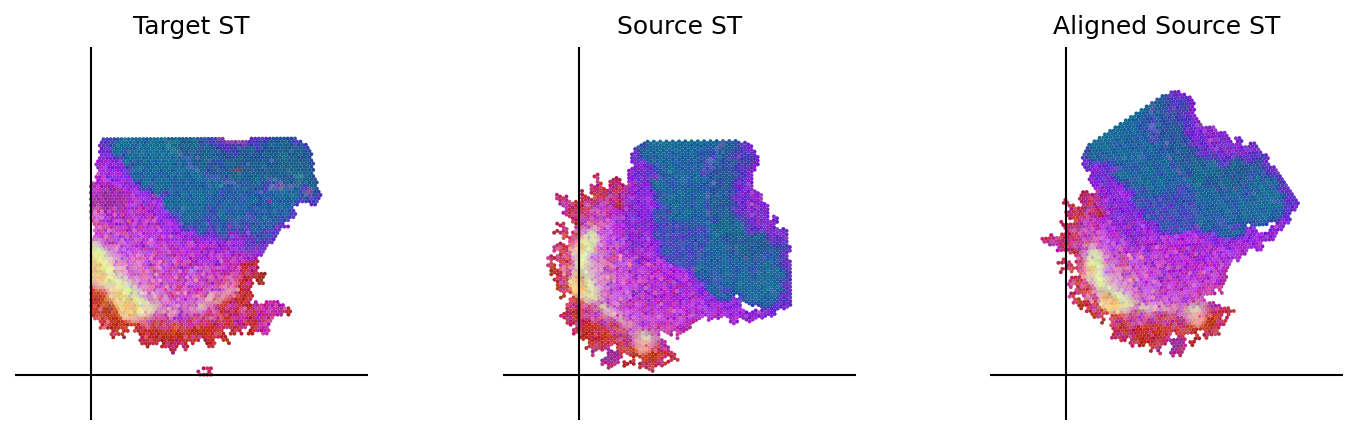
[77]:
loki.plot.show_image(aligned_image)
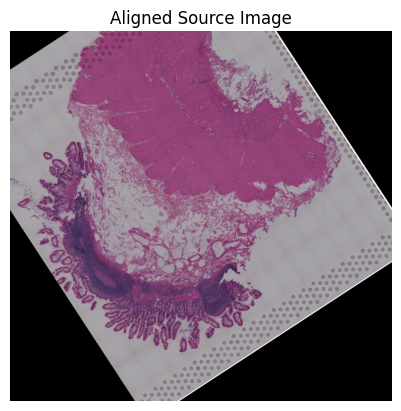
Source7
[78]:
src_sample_name = 'V10F24-048_A1'
ad_path = os.path.join(data_dir, 'visium_data', src_sample_name+'.h5ad')
ad_src, ad_src_coor, src_img = loki.preprocess.prepare_data_for_alignment(ad_path)
Loki Align on ST to ST alignment
Use Loki Align to align ST to ST.
[79]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[80]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
pca_rgb_comb = (pca_comb_features-pca_comb_features.min(axis=0))/(pca_comb_features.max(axis=0)-pca_comb_features.min(axis=0))
pca_rgb_comb[:,[0,1,2]]=pca_rgb_comb[:,[0,2,1]]
pca_hex_comb = [ rgb2hex(pca_rgb_comb[i,:]) for i in range(pca_rgb_comb.shape[0]) ]
[81]:
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[82]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[83]:
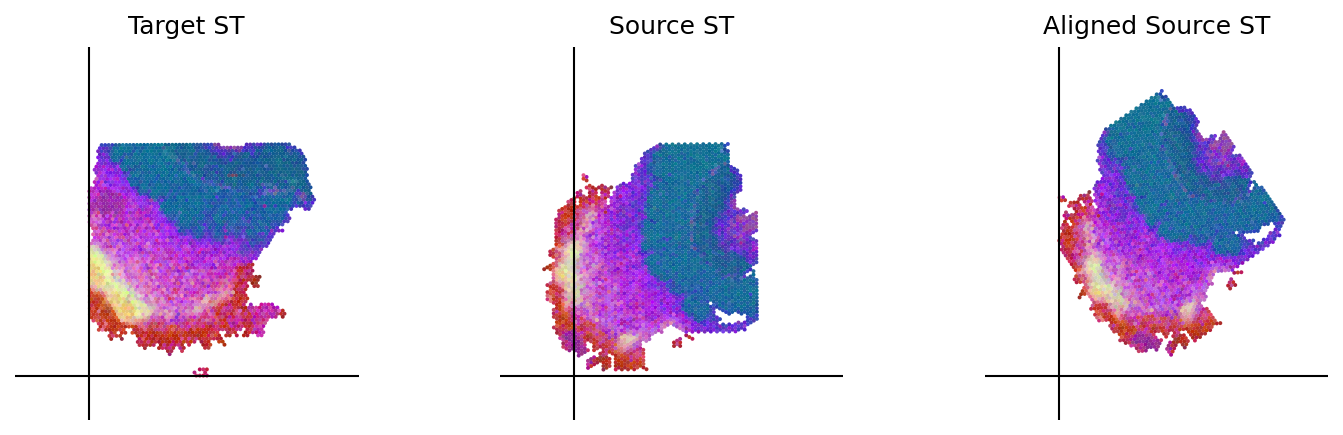
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
[84]:
loki.plot.show_image(aligned_image)
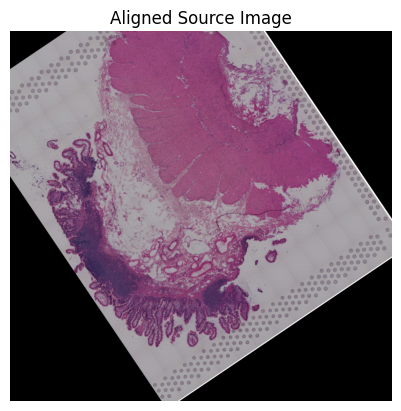
Loki Align on image to ST Alignment
Use Loki Align to align image to ST.
[85]:
tar_features = get_features(tar_sample_name, '_txt_features_zs')
src_features = get_features(src_sample_name, '_txt_features')
[86]:
pca_comb_features, pca_comb_features_batch = loki.utils.get_pca_by_fit(tar_features, src_features)
cpd_coor, homo_coor, aligned_image = loki.align.align_tissue(ad_tar_coor, ad_src_coor, pca_comb_features, src_img)
[87]:
# loki.plot.plot_alignment_with_img(ad_tar_coor, ad_src_coor, homo_coor, tar_img, src_img, aligned_image, pca_hex_comb, tar_features)
[88]:
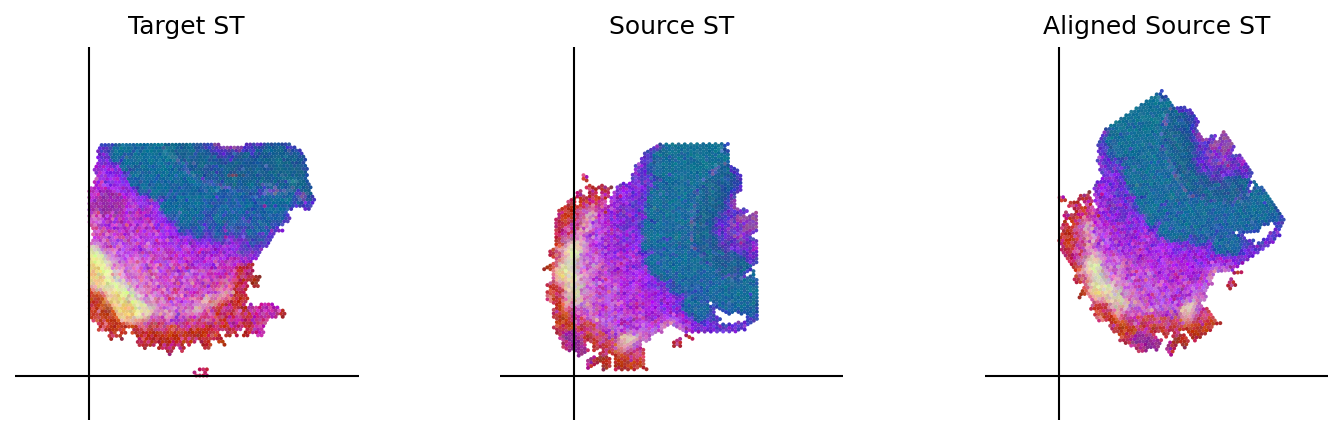
loki.plot.plot_alignment(ad_tar_coor, ad_src_coor, homo_coor, pca_hex_comb, tar_features)
[89]:
loki.plot.show_image(aligned_image)
[ ]: