Loki Annotate - Bulk RNA-seq
This notebook demonstrates how to run Loki Annotate on the bulk RNA-seq dataset. It takes about 2 mins to run this notebook on MacBook Pro.
[1]:
import pandas as pd
import os
import torch
from pathlib import Path
from PIL import Image
import matplotlib.pyplot as plt
import loki.annotate
import loki.preprocess
import loki.utils
import loki.plot
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/timm/models/layers/__init__.py:48: FutureWarning: Importing from timm.models.layers is deprecated, please import via timm.layers
warnings.warn(f"Importing from {__name__} is deprecated, please import via timm.layers", FutureWarning)
We provide the embeddings generated from the OmiCLIP model. The sample data and embeddings are stored in the directory data/loki_annotate/bulk_RNAseq_data, which can be donwloaded from Google Drive link.
Here is a list of the files that are needed to run the tissue annotation on example data:
.
├── adipose
│ ├── checkpoints
│ │ ├── bulk_adipose_txt_features_zeroshot.csv
│ │ ├── samplev02_img_features_zeroshot.csv
│ │ ├── samplev03_img_features_zeroshot.csv
│ │ └── samplev04_img_features_zeroshot.csv
│ └── image_data
│ ├── samplev02_coords.json
│ ├── samplev02_raw.h5ad
│ ├── samplev03_coords.json
│ ├── samplev03_raw.h5ad
│ ├── samplev04_coords.json
│ └── samplev04_raw.h5ad
├── fibroblast
│ ├── checkpoints
│ │ ├── FZ_P14_img_features_zeroshot.csv
│ │ ├── FZ_P18_img_features_zeroshot.csv
│ │ ├── FZ_P20_img_features_zeroshot.csv
│ │ └── bulk_fibroblast_txt_features_zeroshot.csv
│ └── image_data
│ ├── FZ_P14.tif
│ ├── FZ_P14_coords.json
│ ├── FZ_P14_raw.h5ad
│ ├── FZ_P18.tif
│ ├── FZ_P18_coords.json
│ ├── FZ_P18_raw.h5ad
│ ├── FZ_P20.tif
│ ├── FZ_P20_coords.json
│ └── FZ_P20_raw.h5ad
└── tumor
├── checkpoints
│ ├── TCGA-A2-A0CL-01_image_embeddings.pt
│ ├── TCGA-A2-A0CL-01_text_embeddings.pt
│ ├── TCGA-A2-A0CW-01_image_embeddings.pt
│ ├── TCGA-A2-A0CW-01_text_embeddings.pt
│ ├── TCGA-AO-A0J5-01_image_embeddings.pt
│ └── TCGA-AO-A0J5-01_text_embeddings.pt
└── image_data
├── TCGA-A2-A0CL-01_coords.pt
├── TCGA-A2-A0CL-01_downsized.jpg
├── TCGA-A2-A0CW-01_coords.pt
├── TCGA-A2-A0CW-01_downsized.jpg
├── TCGA-AO-A0J5-01_coords.pt
├── TCGA-AO-A0J5-01_downsized.jpg
└── polygons.json
[2]:
data_dir = './data/loki_annotate/bulk_RNAseq_data/'
[3]:
case_name = '_img_features_zeroshot'
Loki Annote with bulk RNA-seq data of tumor
Use Loki Annotate to annotate tumor cell enriched regions in 3 H&E images of breast cancer tissues.
TCGA data
The TCGA breast cancer slide files can be downloaded from the GDC data portal. We provide the image files downsized by 32 times for visualization.
The bulk RNA-seq data can be downloaded from cBioportal.
[ ]:
sample_type = 'tumor'
slide_dict = {
"TCGA-A2-A0CL-01": "TCGA-A2-A0CL-01Z-00-DX1.5342E971-DCD2-42C4-B4FF-E6942A95829E.svs",
"TCGA-A2-A0CW-01": "TCGA-A2-A0CW-01Z-00-DX1.8E313A22-B0E8-44CF-ADEA-8BF29BA23FFE.svs",
"TCGA-AO-A0J5-01": "TCGA-AO-A0J5-01Z-00-DX1.20C14D0C-1A74-4FE9-A5E6-BDDCB8DE7714.svs"
}
[4]:
for case_id, slide_path in slide_dict.items():
print(case_id)
slide_id = Path(slide_path).stem
image_path = os.path.join(data_dir, sample_type, 'image_data', case_id+'_downsized.jpg')
coor_path = os.path.join(data_dir, sample_type, 'image_data', case_id+'_coords.pt')
image_embedding_path = os.path.join(data_dir, sample_type, 'checkpoints', case_id+'_image_embeddings.pt')
omic_embedding_path = os.path.join(data_dir, sample_type, 'checkpoints', case_id+'_text_embeddings.pt')
coor = torch.load(coor_path)
img_features = torch.load(image_embedding_path) # Image embeddings (shape: [n, 768])
bulk_features = torch.load(omic_embedding_path) # Omic features (shape: [768])
weight = loki.annotate.annotate_with_bulk(img_features, bulk_features, tensor=True)[0].squeeze().numpy()
weight = loki.utils.cap_quantile(weight, cap_max=0.99, cap_min=0.1)
polygon_config_path = os.path.join(data_dir, sample_type, 'image_data', 'polygons.json')
polygons, polygons_color, polygons_thickness = loki.utils.read_polygons(polygon_config_path, slide_path)
heatmap, image = loki.plot.plot_heatmap(
coor,
weight,
image_path,
polygons,
polygons_color,
polygons_thickness,
)
loki.plot.show_images_side_by_side(image, heatmap)
TCGA-A2-A0CL-01
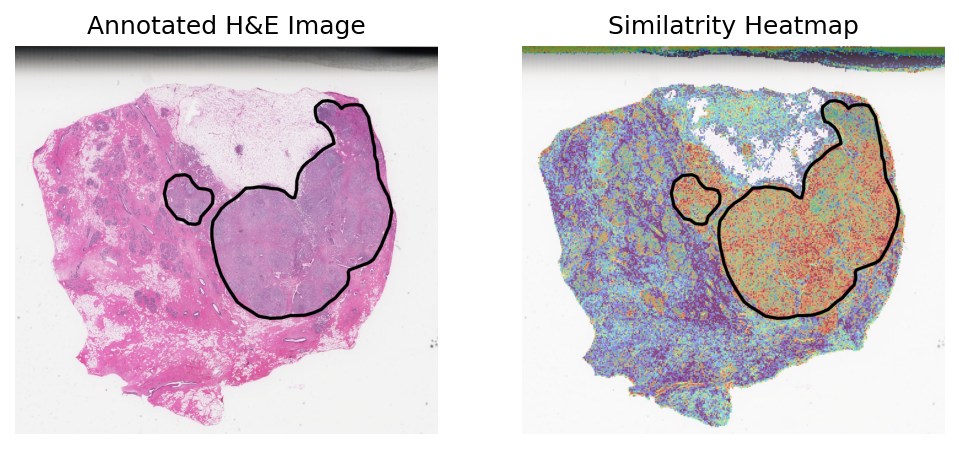
TCGA-A2-A0CW-01
TCGA-AO-A0J5-01
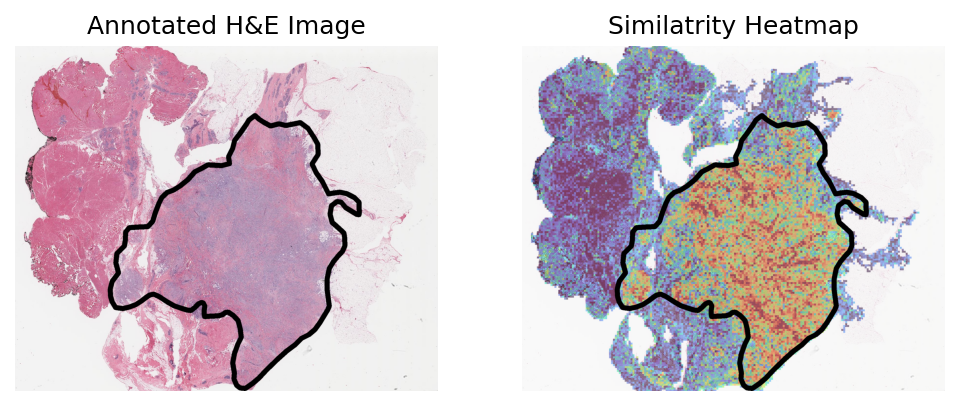
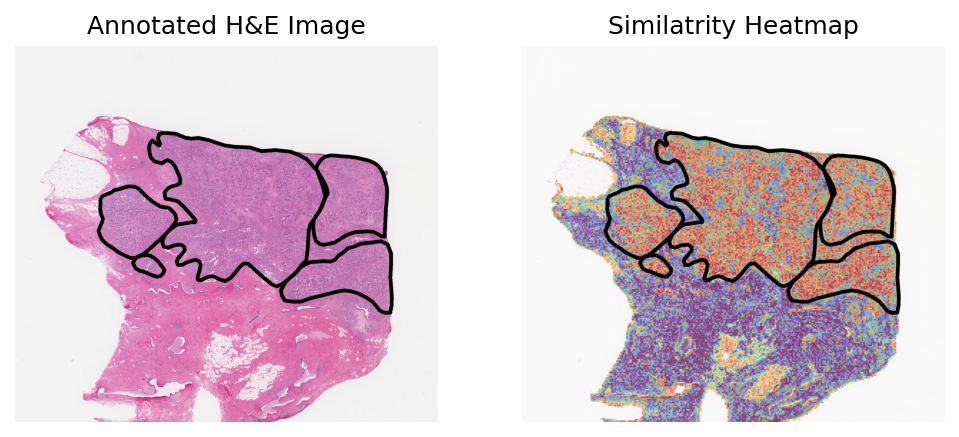
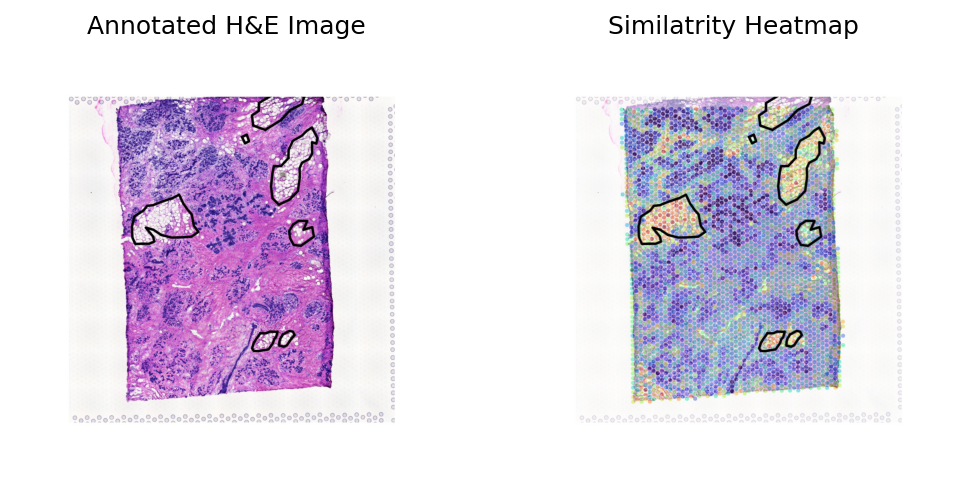
Loki Annote with bulk RNA-seq data of fibroblast
Use Loki Annotate to annotate fibroblast cell enriched regions in 3 H&E images of heart failure tissues.
[4]:
sample_type = 'fibroblast'
s=35
linewidth=5
xlim=[0,10000]
ylim=[0,10000]
bulk_text_feature = pd.read_csv(os.path.join(data_dir, sample_type, 'checkpoints', 'bulk_fibroblast_txt_features_zeroshot.csv'), index_col=0)
Heart failure sample 1
[5]:
sample_name = 'FZ_P20'
st_data_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_raw.h5ad')
json_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_coords.json')
st_ad, library_id, roi_polygon = loki.preprocess.load_data_for_annotation(st_data_path, json_path)
fullres_img_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'.tif')
fullres_img = Image.open(fullres_img_path)
img_feature_path = os.path.join(data_dir, sample_type, 'checkpoints', sample_name+case_name+'.csv')
img_feature = pd.read_csv(img_feature_path, index_col=0)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/loki/preprocess.py:282: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
st_ad.obs[["pixel_y", "pixel_x"]] = None # Ensure the columns exist
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/PIL/Image.py:3368: DecompressionBombWarning: Image size (93006507 pixels) exceeds limit of 89478485 pixels, could be decompression bomb DOS attack.
warnings.warn(
[6]:
loki.plot.plot_img_with_annotation(fullres_img, roi_polygon, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg'))
plt.close()
[7]:
st_ad.obs['bulk_simi'] = loki.annotate.annotate_with_bulk(img_feature, bulk_text_feature)
[8]:
loki.plot.plot_annotation_heatmap(st_ad, roi_polygon, s, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg'))
plt.close()
[9]:
image_path = os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg')
image = loki.annotate.load_image_annotation(image_path)
heatmap_path = os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg')
heatmap = loki.annotate.load_image_annotation(heatmap_path)
heatmap = loki.plot.blend_images(heatmap, image)
[10]:
loki.plot.show_images_side_by_side(image, heatmap)
Heart failure sample 2
[11]:
sample_name = 'FZ_P18'
st_data_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_raw.h5ad')
json_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_coords.json')
st_ad, library_id, roi_polygon = loki.preprocess.load_data_for_annotation(st_data_path, json_path)
fullres_img_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'.tif')
fullres_img = Image.open(fullres_img_path)
img_feature_path = os.path.join(data_dir, sample_type, 'checkpoints', sample_name+case_name+'.csv')
img_feature = pd.read_csv(img_feature_path, index_col=0)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/anndata/_core/anndata.py:1756: UserWarning: Variable names are not unique. To make them unique, call `.var_names_make_unique`.
utils.warn_names_duplicates("var")
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/loki/preprocess.py:282: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
st_ad.obs[["pixel_y", "pixel_x"]] = None # Ensure the columns exist
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/PIL/Image.py:3368: DecompressionBombWarning: Image size (96294069 pixels) exceeds limit of 89478485 pixels, could be decompression bomb DOS attack.
warnings.warn(
[12]:
loki.plot.plot_img_with_annotation(fullres_img, roi_polygon, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg'))
plt.close()
[13]:
st_ad.obs['bulk_simi'] = loki.annotate.annotate_with_bulk(img_feature, bulk_text_feature)
[14]:
loki.plot.plot_annotation_heatmap(st_ad, roi_polygon, s, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg'))
plt.close()
[15]:
image_path = os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg')
image = loki.annotate.load_image_annotation(image_path)
heatmap_path = os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg')
heatmap = loki.annotate.load_image_annotation(heatmap_path)
heatmap = loki.plot.blend_images(heatmap, image)
[16]:
loki.plot.show_images_side_by_side(image, heatmap)
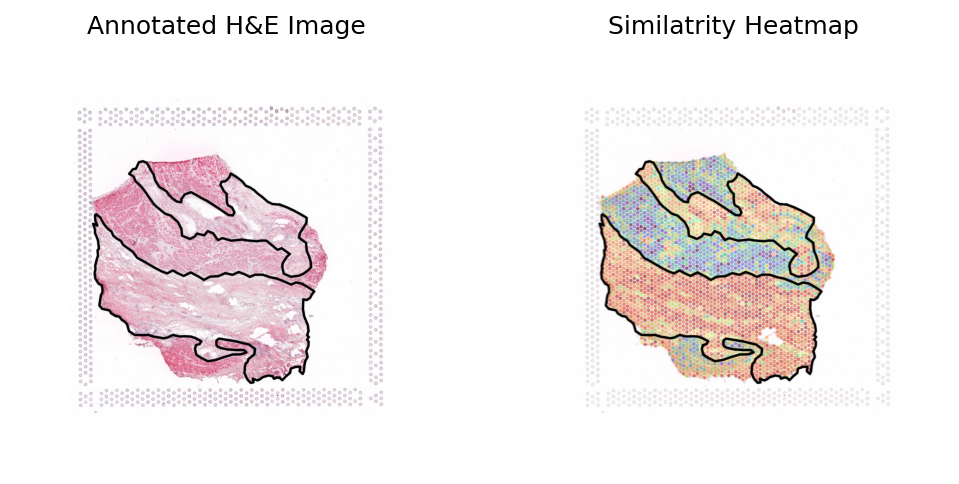
Heart failure sample 3
[17]:
sample_name = 'FZ_P14'
st_data_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_raw.h5ad')
json_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_coords.json')
st_ad, library_id, roi_polygon = loki.preprocess.load_data_for_annotation(st_data_path, json_path, in_tissue=False)
fullres_img_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'.tif')
fullres_img = Image.open(fullres_img_path)
img_feature_path = os.path.join(data_dir, sample_type, 'checkpoints', sample_name+case_name+'.csv')
img_feature = pd.read_csv(img_feature_path, index_col=0)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/PIL/Image.py:3368: DecompressionBombWarning: Image size (94551858 pixels) exceeds limit of 89478485 pixels, could be decompression bomb DOS attack.
warnings.warn(
[18]:
loki.plot.plot_img_with_annotation(fullres_img, roi_polygon, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg'))
plt.close()
[19]:
st_ad.obs['bulk_simi'] = loki.annotate.annotate_with_bulk(img_feature, bulk_text_feature)
[20]:
loki.plot.plot_annotation_heatmap(st_ad, roi_polygon, s, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg'))
plt.close()
[21]:
image_path = os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg')
image = loki.annotate.load_image_annotation(image_path)
heatmap_path = os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg')
heatmap = loki.annotate.load_image_annotation(heatmap_path)
heatmap = loki.plot.blend_images(heatmap, image)
[22]:
loki.plot.show_images_side_by_side(image, heatmap)
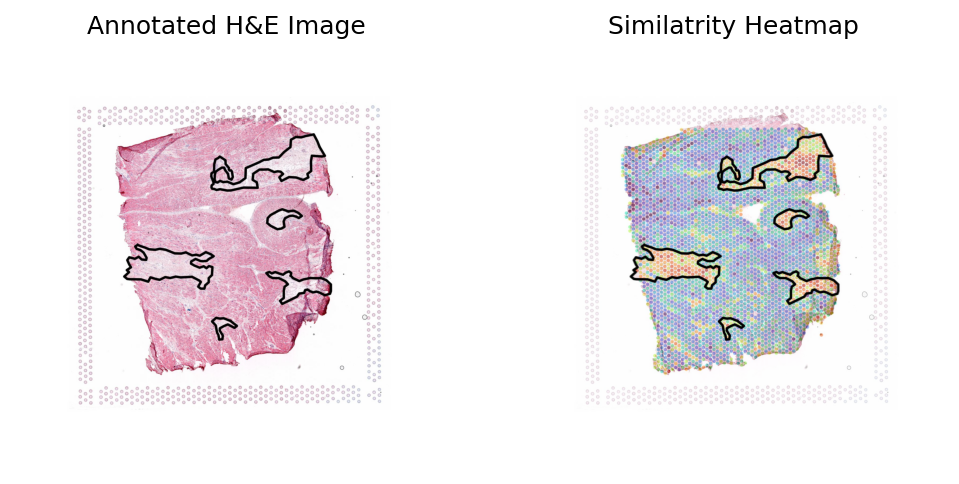
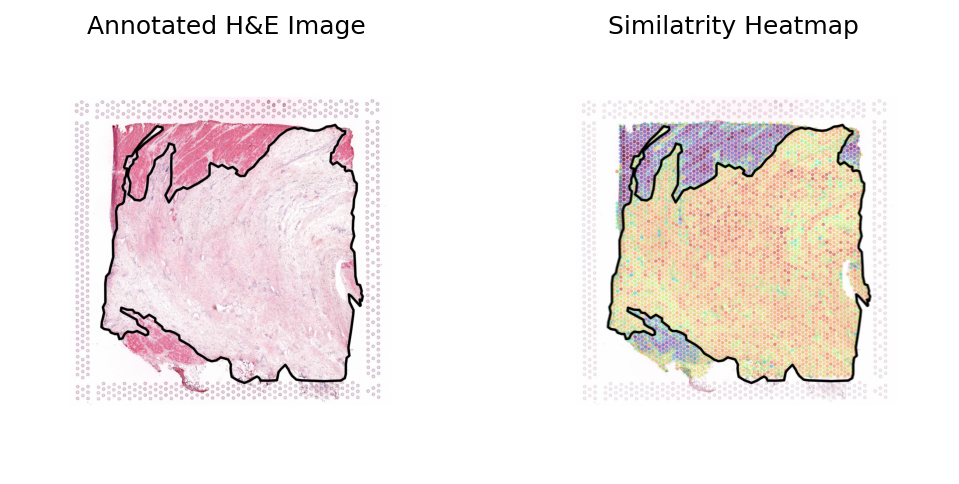
Loki Annote with bulk RNA-seq data of adipose
Use Loki Annotate to annotate adipose cell enriched regions in 3 H&E images of normal breast tissues.
[23]:
sample_type = 'adipose'
bulk_text_feature = pd.read_csv(os.path.join(data_dir, sample_type, 'checkpoints', 'bulk_adipose_txt_features_zeroshot.csv'), index_col=0)
Breast sample 1
[24]:
sample_name = 'samplev02'
st_data_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_raw.h5ad')
json_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_coords.json')
st_ad, library_id, roi_polygon = loki.preprocess.load_data_for_annotation(st_data_path, json_path)
fullres_img = st_ad.uns['spatial'][library_id]['images']['fullres']
img_feature_path = os.path.join(data_dir, sample_type, 'checkpoints', sample_name+case_name+'.csv')
img_feature = pd.read_csv(img_feature_path, index_col=0)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/loki/preprocess.py:282: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
st_ad.obs[["pixel_y", "pixel_x"]] = None # Ensure the columns exist
[25]:
s=50
linewidth=4
xlim=[0,4000]
ylim=[0,4000]
loki.plot.plot_img_with_annotation(fullres_img, roi_polygon, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg'))
plt.close()
[26]:
st_ad.obs['bulk_simi'] = loki.annotate.annotate_with_bulk(img_feature, bulk_text_feature)
[27]:
loki.plot.plot_annotation_heatmap(st_ad, roi_polygon, s, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg'))
plt.close()
[28]:
image_path = os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg')
image = loki.annotate.load_image_annotation(image_path)
heatmap_path = os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg')
heatmap = loki.annotate.load_image_annotation(heatmap_path)
heatmap = loki.plot.blend_images(heatmap, image)
[29]:
loki.plot.show_images_side_by_side(image, heatmap)
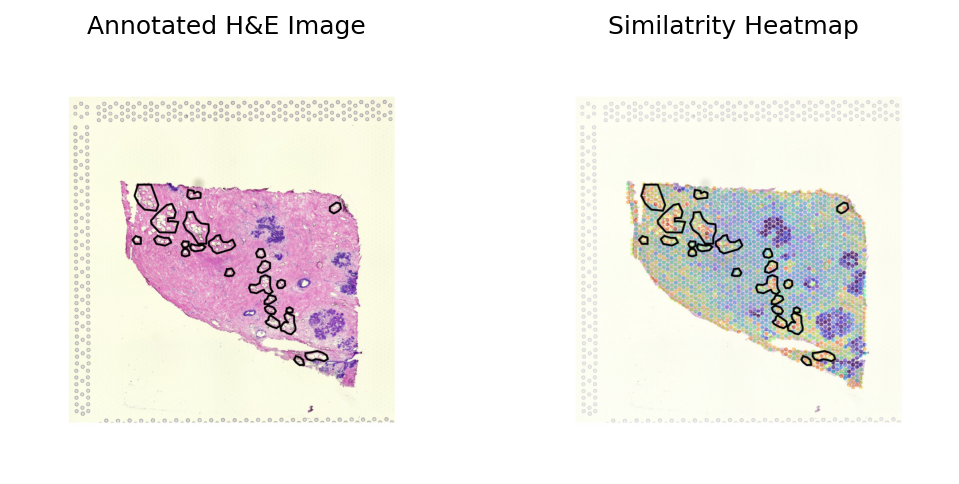
Breast sample 2
[30]:
sample_name = 'samplev03'
st_data_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_raw.h5ad')
json_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_coords.json')
st_ad, library_id, roi_polygon = loki.preprocess.load_data_for_annotation(st_data_path, json_path)
fullres_img = st_ad.uns['spatial'][library_id]['images']['fullres']
img_feature_path = os.path.join(data_dir, sample_type, 'checkpoints', sample_name+case_name+'.csv')
img_feature = pd.read_csv(img_feature_path, index_col=0)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/loki/preprocess.py:282: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
st_ad.obs[["pixel_y", "pixel_x"]] = None # Ensure the columns exist
[31]:
s=55
linewidth=4
xlim=[1500,21000]
ylim=[1500,21000]
loki.plot.plot_img_with_annotation(fullres_img, roi_polygon, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg'))
plt.close()
[32]:
st_ad.obs['bulk_simi'] = loki.annotate.annotate_with_bulk(img_feature, bulk_text_feature)
[33]:
loki.plot.plot_annotation_heatmap(st_ad, roi_polygon, s, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg'))
plt.close()
[34]:
image_path = os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg')
image = loki.annotate.load_image_annotation(image_path)
heatmap_path = os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg')
heatmap = loki.annotate.load_image_annotation(heatmap_path)
heatmap = loki.plot.blend_images(heatmap, image)
[35]:
loki.plot.show_images_side_by_side(image, heatmap)
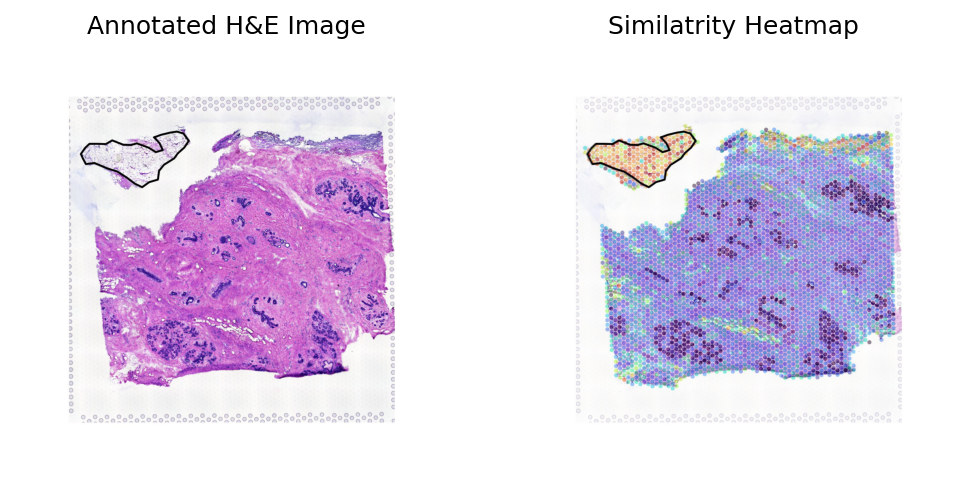
Breast sample 3
[36]:
sample_name = 'samplev04'
st_data_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_raw.h5ad')
json_path = os.path.join(data_dir, sample_type, 'image_data', sample_name+'_coords.json')
st_ad, library_id, roi_polygon = loki.preprocess.load_data_for_annotation(st_data_path, json_path)
fullres_img = st_ad.uns['spatial'][library_id]['images']['fullres']
img_feature_path = os.path.join(data_dir, sample_type, 'checkpoints', sample_name+case_name+'.csv')
img_feature = pd.read_csv(img_feature_path, index_col=0)
/opt/anaconda3/envs/loki/lib/python3.9/site-packages/loki/preprocess.py:282: ImplicitModificationWarning: Trying to modify attribute `.obs` of view, initializing view as actual.
st_ad.obs[["pixel_y", "pixel_x"]] = None # Ensure the columns exist
[37]:
s=55
linewidth=5
xlim=[2000,21000]
ylim=[2000,21000]
loki.plot.plot_img_with_annotation(fullres_img, roi_polygon, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg'))
plt.close()
[38]:
st_ad.obs['bulk_simi'] = loki.annotate.annotate_with_bulk(img_feature, bulk_text_feature)
[39]:
loki.plot.plot_annotation_heatmap(st_ad, roi_polygon, s, linewidth, xlim, ylim)
plt.savefig(os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg'))
plt.close()
[40]:
image_path = os.path.join(data_dir, sample_type, sample_name+'_fullres.jpg')
image = loki.annotate.load_image_annotation(image_path)
heatmap_path = os.path.join(data_dir, sample_type, sample_name+'_heatmap.jpg')
heatmap = loki.annotate.load_image_annotation(heatmap_path)
heatmap = loki.plot.blend_images(heatmap, image)
[41]:
loki.plot.show_images_side_by_side(image, heatmap)
[ ]: