cellDancer - Estimating Cell-dependent RNA Velocity¶
cellDancer is a modularized, parallelized, and scalable tool based on a deep learning framework for the RNA velocity analysis of scRNA-seq proposed by Li et al. (Nature Biotechnology, 2023). RNA velocity reflects the dynamic process of cell state transitions based on scRNA-seq experiment data. cellDancer enables the prediction of RNA velocity and the dynamic kinetics of RNA in single-cell resolution.
cellDancer’s key applications¶
Enable accurate inference of dynamic cell state transitions in heterogeneous cell populations.
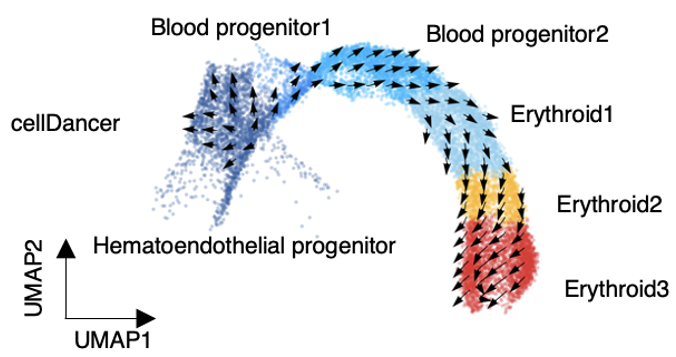
Example: Mouse gastrulation erythroid maturation.
Estimate cell-specific transcription (α), splicing (β) and degradation (γ) rates for each gene and reveal RNA turnover strategies.
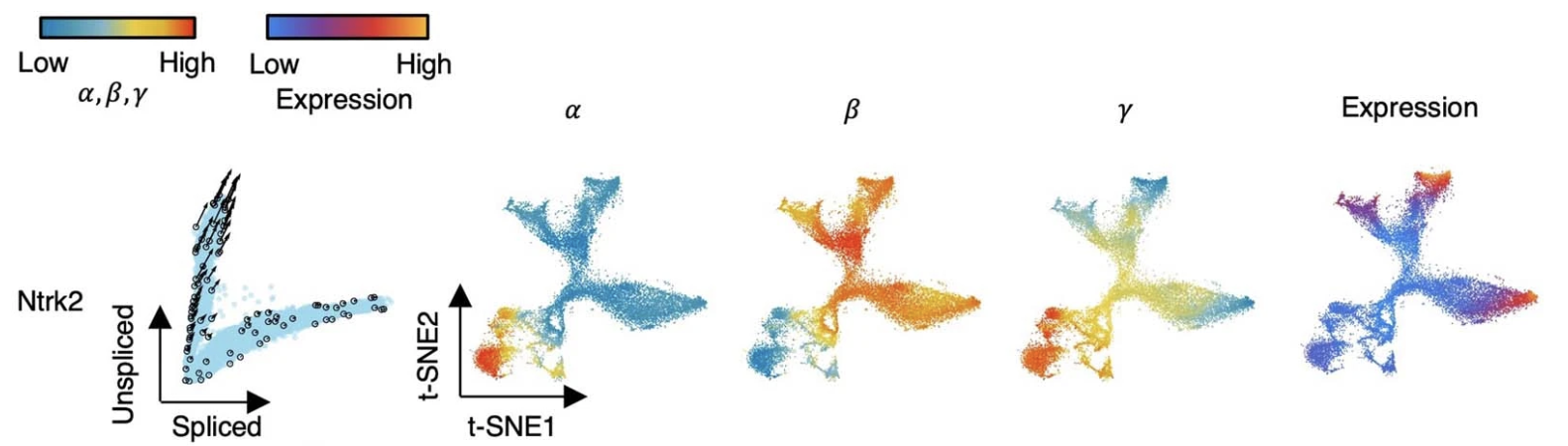
Example: Mouse hippocampus development.
Improves downstream analysis such as vector field predictions.
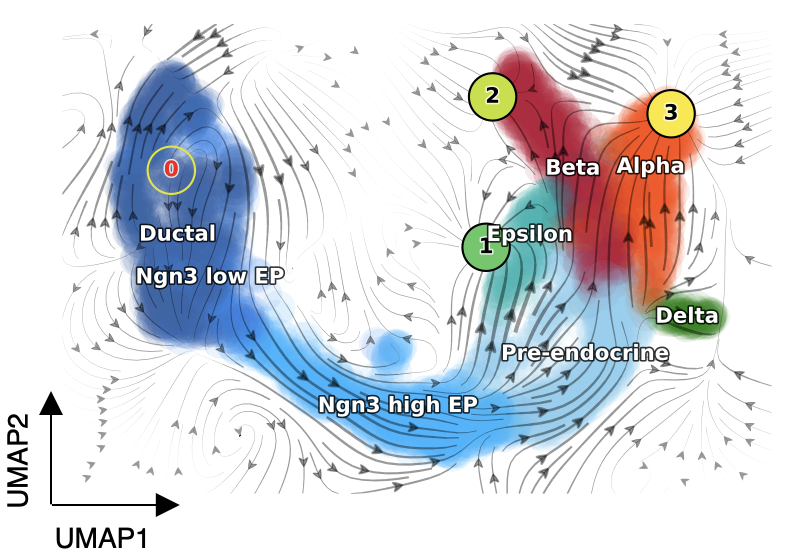
Example: Mouse pancreatic endocrinogenesis.
Latest news¶
Our work of cellDancer has been published at Nature Biotechnology! (4/3/2023)
cellDancer has been released to PyPI (3/21/2023).
Support¶
Welcome bug reports and suggestions to our Github issue page!
Table of contents¶
cellDancer
- About cellDancer
- API
- celldancer.adata_to_df_with_embed
- celldancer.to_dynamo
- celldancer.export_velocity_to_dynamo
- celldancer.velocity
- celldancer.compute_cell_velocity
- celldancer.pseudo_time
- celldancer.embedding_kinetic_para
- celldancer.cdplt.scatter_gene
- celldancer.cdplt.scatter_cell
- celldancer.cdplt.plot_kinetic_para
- celldancer.cdplt.PTO_Graph
- celldancer.simulation.simulate
- Release Notes
- Frequently asked questions
Tutorials
- Installation
- Data Preparation
- Case study 1: Gastrulation erythroid maturation
- Case Study 2: Mouse hippocampus development
- Import packages
- Load data
- Estimate RNA velocity for sample genes
- Visualize the phase portraits
- Project the RNA velocity onto the embedding space
- Estimate pseudotime
- Display the abundance of spliced RNA along pseudotime
- Visualize the reaction rates, the abundance of unspliced RNA, and the abundance of spliced RNA on embedding space
- Case study 3: Pancreatic endocrinogenesis
- Case study 4: Human embryo glutamatergic neurogenesis